A Motif in the F Homomorph of Rabbit Haemorrhagic Disease Virus Polymerase Is Important for the Subcellular Localisation of the Protein and Its Ability to Induce Redistribution of Golgi Membranes
- PMID: 28763035
- PMCID: PMC5580459
- DOI: 10.3390/v9080202
A Motif in the F Homomorph of Rabbit Haemorrhagic Disease Virus Polymerase Is Important for the Subcellular Localisation of the Protein and Its Ability to Induce Redistribution of Golgi Membranes
Abstract
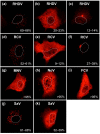
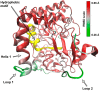
Rabbit haemorrhagic disease virus (RHDV) is a calicivirus that infects and frequently kills rabbits. Previously, we showed that the RHDV RNA-dependent RNA polymerase (RdRp) is associated with distinct, but yet uncharacterised subcellular structures and is capable of inducing a redistribution of Golgi membranes. In this study, we identified a partially hidden hydrophobic motif that determines the subcellular localisation of recombinant RHDV RdRp in transfected cells. This novel motif, 189LLWGCDVGVAVCAAAVFHNICY210, is located within the F homomorph, between the conserved F3 and A motifs of the core RdRp domain. Amino acid substitutions that decrease the hydrophobicity of this motif reduced the ability of the protein to accumulate in multiple subcellular foci and to induce a rearrangement of the Golgi network. Furthermore, preliminary molecular dynamics simulations suggest that the RHDV RdRp could align with the negatively charged surfaces of biological membranes and undergo a conformational change involving the F homomorph. These changes would expose the newly identified hydrophobic motif so it could immerse itself into the outer leaflet of intracellular membranes.
Keywords: Golgi membranes; RHDV; RNA-dependent RNA polymerase; Rabbit haemorrhagic disease virus; caliciviruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Calicivirus RNA-Dependent RNA Polymerases: Evolution, Structure, Protein Dynamics, and Function.Front Microbiol. 2019 Jun 6;10:1280. doi: 10.3389/fmicb.2019.01280. eCollection 2019. Front Microbiol. 2019. PMID: 31244803 Free PMC article. Review.
-
RNA-Dependent RNA Polymerases of Both Virulent and Benign Rabbit Caliciviruses Induce Striking Rearrangement of Golgi Membranes.PLoS One. 2017 Jan 10;12(1):e0169913. doi: 10.1371/journal.pone.0169913. eCollection 2017. PLoS One. 2017. PMID: 28072826 Free PMC article.
-
Purification and Biochemical Characterisation of Rabbit Calicivirus RNA-Dependent RNA Polymerases and Identification of Non-Nucleoside Inhibitors.Viruses. 2016 Apr 14;8(4):100. doi: 10.3390/v8040100. Viruses. 2016. PMID: 27089358 Free PMC article.
-
Expression and partial characterisation of rabbit haemorrhagic disease virus non-structural proteins.Virology. 2015 Oct;484:69-79. doi: 10.1016/j.virol.2015.05.004. Epub 2015 Jun 11. Virology. 2015. PMID: 26071926
-
Genetic variation and phylogenetic analysis of rabbit haemorrhagic disease virus (RHDV) strains.Acta Biochim Pol. 2012;59(4):459-65. Epub 2012 Dec 13. Acta Biochim Pol. 2012. PMID: 23240105 Review.
Cited by
-
Structure and Function of Caliciviral RNA Polymerases.Viruses. 2017 Nov 6;9(11):329. doi: 10.3390/v9110329. Viruses. 2017. PMID: 29113097 Free PMC article. Review.
-
Immune Response Modulation by Caliciviruses.Front Immunol. 2019 Oct 1;10:2334. doi: 10.3389/fimmu.2019.02334. eCollection 2019. Front Immunol. 2019. PMID: 31632406 Free PMC article. Review.
-
Rabbit Hemorrhagic Disease Virus Isolated from Diseased Alpine Musk Deer (Moschussifanicus).Viruses. 2020 Aug 17;12(8):897. doi: 10.3390/v12080897. Viruses. 2020. PMID: 32824417 Free PMC article.
-
Calicivirus RNA-Dependent RNA Polymerases: Evolution, Structure, Protein Dynamics, and Function.Front Microbiol. 2019 Jun 6;10:1280. doi: 10.3389/fmicb.2019.01280. eCollection 2019. Front Microbiol. 2019. PMID: 31244803 Free PMC article. Review.
-
Nucleolin interacts with the rabbit hemorrhagic disease virus replicase RdRp, nonstructural proteins p16 and p23, playing a role in virus replication.Virol Sin. 2022 Feb;37(1):48-59. doi: 10.1016/j.virs.2022.01.004. Epub 2022 Jan 12. Virol Sin. 2022. PMID: 35234629 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

