MicroRNA expression profiles differ between primary myofiber of lean and obese pig breeds
- PMID: 28759650
- PMCID: PMC5536276
- DOI: 10.1371/journal.pone.0181897
MicroRNA expression profiles differ between primary myofiber of lean and obese pig breeds
Abstract
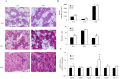
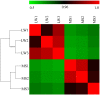
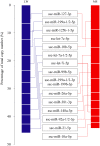
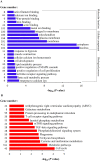
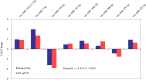
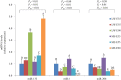
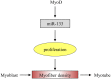
MicroRNAs (miRNAs) are non-coding small miRNAs ~22 nucleotides in length and play a vital role in muscle development by binding to messenger RNAs (mRNAs). Large White (LW, a lean type pig) and Meishan pigs (MS, a Chinese indigenous obese breed) have significant postnatal phenotype differences in growth rate, muscle mass and meat quality, and these differences are programmed during prenatal muscle development. Little research shed light directly on the miRNA transcriptome difference in prenatal muscles between these two distinct pig breeds. Myofiber phenotypes of LW and MS were measured at developmental stages of 35, 55 and 90 days post-conception (dpc), which revealed that the myogenesis process is more intense in MS than in LW at 35 dpc. To investigate the role of miRNAs involved in regulating muscle development at earlier stages of myogenesis and decipher the miRNAs transcriptome difference between LW and MS, here, the miRNAomes of longissimus dorsi muscle collected at 35 dpc from female LW and MS were analyzed by deep sequencing. Overall, 1147 unique miRNAs comprising 434 known miRNAs, 239 conserved miRNAs and 474 candidate miRNAs were identified. Expression analysis of the 10 most abundant miRNAs in every library indicated that functional miRNAome may be a small amount and tend to be greater expressed. These sets of miRNA may play house keeping roles that were involved in myogenesis. A total of 87 miRNAs were significantly differentially expressed between LW and MS (reads > 1000, P < 0.05). Gene ontology (GO) and KEGG pathway enrichment analysis revealed that the differentially expressed miRNAs (DE miRNAs) were associated mainly with muscle contraction, WNT, mTOR, and MAPK signaling pathways. Some myogenesis related miRNAs (miR-133, miR-1, miR-206 and miR-148a) are highly abundant in MS, while other miRNAs (let-7 family, miR-214, miR-181) highly expressed in LW. In addition, the expression patterns of miRNAs (miR-1, -133, -206) at three prenatal stages (35, 55 and 90 dpc) were determined using qRT-PCR. Notably, ssc-miR-133 was significantly more highly expressed in LW pigs skeletal muscle at all prenatal stages compared with its expression in LW pigs skeletal muscle. Taken together, the main functional miRNAs during muscle development are different between lean and obese pig breeds. The present study adds new information to existing data on porcine miRNAs and will be helpful to investigate the dominant (main functional) muscle-related miRNAs sets in different pig breeds.
Conflict of interest statement
Figures


Similar articles
-
Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue.PLoS One. 2015 Oct 27;10(10):e0141138. doi: 10.1371/journal.pone.0141138. eCollection 2015. PLoS One. 2015. PMID: 26505482 Free PMC article.
-
Comparison of skeletal muscle miRNA and mRNA profiles among three pig breeds.Mol Genet Genomics. 2016 Apr;291(2):559-73. doi: 10.1007/s00438-015-1126-3. Epub 2015 Oct 12. Mol Genet Genomics. 2016. PMID: 26458558
-
Comparative Analyses between Skeletal Muscle miRNAomes from Large White and Min Pigs Revealed MicroRNAs Associated with Postnatal Muscle Hypertrophy.PLoS One. 2016 Jun 2;11(6):e0156780. doi: 10.1371/journal.pone.0156780. eCollection 2016. PLoS One. 2016. PMID: 27253583 Free PMC article.
-
Effects of microRNAs on skeletal muscle development.Gene. 2018 Aug 20;668:107-113. doi: 10.1016/j.gene.2018.05.039. Epub 2018 May 25. Gene. 2018. PMID: 29775754 Review.
-
MicroRNA in Skeletal Muscle: Its Crucial Roles in Signal Proteins, Mus cle Fiber Type, and Muscle Protein Synthesis.Curr Protein Pept Sci. 2017;18(6):579-588. doi: 10.2174/1389203717666160621122405. Curr Protein Pept Sci. 2017. PMID: 27341987 Review.
Cited by
-
Integrated miRNA-mRNA transcriptomic analysis reveals epigenetic-mediated embryonic muscle growth differences between Wuzhishan and Landrace pigs1.J Anim Sci. 2019 Apr 29;97(5):1967-1978. doi: 10.1093/jas/skz091. J Anim Sci. 2019. PMID: 31222274 Free PMC article.
-
A Comparative Analysis of Metabolic Profiles of Embryonic Skeletal Muscle from Lantang and Landrace Pigs.Animals (Basel). 2022 Feb 10;12(4):420. doi: 10.3390/ani12040420. Animals (Basel). 2022. PMID: 35203128 Free PMC article.
-
Effects of exogenous α-oxoglutarate on proline accumulation, ammonium assimilation and photosynthesis of soybean seedling (Glycine max(L.) Merr.) exposed to cold stress.Sci Rep. 2020 Oct 12;10(1):17017. doi: 10.1038/s41598-020-74094-w. Sci Rep. 2020. PMID: 33046814 Free PMC article.
-
LncRNA IMFlnc1 promotes porcine intramuscular adipocyte adipogenesis by sponging miR-199a-5p to up-regulate CAV-1.BMC Mol Cell Biol. 2020 Nov 4;21(1):77. doi: 10.1186/s12860-020-00324-8. BMC Mol Cell Biol. 2020. PMID: 33148167 Free PMC article.
-
Genetic regulation and variation of expression of miRNA and mRNA transcripts in fetal muscle tissue in the context of sex, dam and variable fetal weight.Biol Sex Differ. 2022 May 12;13(1):24. doi: 10.1186/s13293-022-00433-3. Biol Sex Differ. 2022. PMID: 35550009 Free PMC article.
References
-
- Lefaucheur L, Ecolan P, Plantard L, Gueguen N. New insights into muscle fiber types in the pig. J Histochem Cytochem. 2002;50(5):719–30. doi: 10.1177/002215540205000513 - DOI - PubMed
-
- Lefaucheur L, Ecolan P. Pattern of muscle fiber formation in Large White and Meishan pigs. Arch Tierz Dummerstorf. 2005;48(Special):117–22. CABI:20053073927
-
- Xu YJ, Jin ML, Wang LJ, Zhang AD, Zuo B, Xu DQ, et al. Differential proteome analysis of porcine skeletal muscles between Meishan and Large White. J Anim Sci. 2009;87(8):2519–27. doi: 10.2527/jas.2008-1708 - DOI - PubMed
-
- Choe JH, Choi YM, Lee SH, Shin HG, Ryu YC, Hong KC, et al. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci. 2008;80(2):355–62. doi: 10.1016/j.meatsci.2007.12.019 . - DOI - PubMed
-
- Maltin CA, Sinclair KD, Warriss PD, Grant CM, Porter AD, Delday MI, et al. The effects of age at slaughter, genotype and finishing system on the biochemical properties, muscle fibre type characteristics and eating quality of bull beef from suckled calves. Anim Sci. 1998;66:341–8 doi: 10.1017/S1357729800009450 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

