Integrating RNA sequencing into neuro-oncology practice
- PMID: 28746860
- PMCID: PMC5659901
- DOI: 10.1016/j.trsl.2017.06.013
Integrating RNA sequencing into neuro-oncology practice
Abstract
Malignant tumors of the central nervous system (CNS) cause substantial morbidity and mortality, yet efforts to optimize chemo- and radiotherapy have largely failed to improve dismal prognoses. Over the past decade, RNA sequencing (RNA-seq) has emerged as a powerful tool to comprehensively characterize the transcriptome of CNS tumor cells in one high-throughput step, leading to improved understanding of CNS tumor biology and suggesting new routes for targeted therapies. RNA-seq has been instrumental in improving the diagnostic classification of brain tumors, characterizing oncogenic fusion genes, and shedding light on intratumor heterogeneity. Currently, RNA-seq is beginning to be incorporated into regular neuro-oncology practice in the form of precision neuro-oncology programs, which use information from tumor sequencing to guide implementation of personalized targeted therapies. These programs show great promise in improving patient outcomes for tumors where single agent trials have been ineffective. As RNA-seq is a relatively new technique, many further applications yielding new advances in CNS tumor research and management are expected in the coming years.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
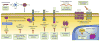

Similar articles
-
RNA-seq Fusion Detection in Clinical Oncology.Adv Exp Med Biol. 2022;1361:163-175. doi: 10.1007/978-3-030-91836-1_9. Adv Exp Med Biol. 2022. PMID: 35230688
-
Development of the CNS TAP tool for the selection of precision medicine therapies in neuro-oncology.J Neurooncol. 2018 Mar;137(1):155-169. doi: 10.1007/s11060-017-2708-1. Epub 2017 Dec 12. J Neurooncol. 2018. PMID: 29235051 Free PMC article.
-
Foundations of Neuro-Oncology: A Multidisciplinary Approach.World Neurosurg. 2021 Jul;151:392-401. doi: 10.1016/j.wneu.2021.02.059. Epub 2021 Feb 20. World Neurosurg. 2021. PMID: 33618043 Review.
-
"Zooming in" on Glioblastoma: Understanding Tumor Heterogeneity and its Clinical Implications in the Era of Single-Cell Ribonucleic Acid Sequencing.Neurosurgery. 2021 Feb 16;88(3):477-486. doi: 10.1093/neuros/nyaa305. Neurosurgery. 2021. PMID: 32674143 Review.
-
Systematic discovery of gene fusions in pediatric cancer by integrating RNA-seq and WGS.BMC Cancer. 2023 Jul 3;23(1):618. doi: 10.1186/s12885-023-11054-3. BMC Cancer. 2023. PMID: 37400763 Free PMC article.
Cited by
-
Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue.BMC Med Genomics. 2019 Jun 17;12(1):86. doi: 10.1186/s12920-019-0538-z. BMC Med Genomics. 2019. PMID: 31208411 Free PMC article.
-
Therapy of breast cancer brain metastases: challenges, emerging treatments and perspectives.Ther Adv Med Oncol. 2018 Jun 22;10:1758835918780312. doi: 10.1177/1758835918780312. eCollection 2018. Ther Adv Med Oncol. 2018. PMID: 29977353 Free PMC article. Review.
-
Molecular tools for the pathologic diagnosis of central nervous system tumors.Neurooncol Pract. 2019 Jan;6(1):4-16. doi: 10.1093/nop/npy041. Epub 2018 Nov 5. Neurooncol Pract. 2019. PMID: 31386041 Free PMC article. Review.
-
Metabolic modeling-based drug repurposing in Glioblastoma.Sci Rep. 2022 Jul 1;12(1):11189. doi: 10.1038/s41598-022-14721-w. Sci Rep. 2022. PMID: 35778411 Free PMC article.
-
A Toolkit for Profiling the Immune Landscape of Pediatric Central Nervous System Malignancies.Front Immunol. 2022 Apr 7;13:864423. doi: 10.3389/fimmu.2022.864423. eCollection 2022. Front Immunol. 2022. PMID: 35464481 Free PMC article. Review.
References
-
- Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 144(5):646–74. - PubMed
-
- Sager M, Yeat NC, Pajaro-Van der Stadt S, Lin C, Ren Q, Lin J. Transcriptomics in cancer diagnostics: developments in technology, clinical research and commercialization. Expert Review of Molecular Diagnostics. 2015;15(12):1589–603. - PubMed
-
- van der Burg SH, Arens R, Ossendorp F, van Hall T, Melief CJ. Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat Rev Cancer. 2016;16(4):219–33. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

