Outer nuclear membrane protein Kuduk modulates the LINC complex and nuclear envelope architecture
- PMID: 28716842
- PMCID: PMC5584142
- DOI: 10.1083/jcb.201606043
Outer nuclear membrane protein Kuduk modulates the LINC complex and nuclear envelope architecture
Abstract
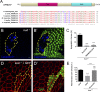
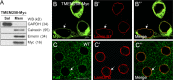
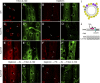
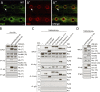
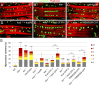
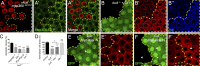
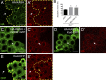
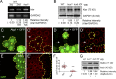
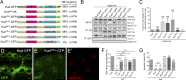
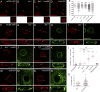
Linker of nucleoskeleton and cytoskeleton (LINC) complexes spanning the nuclear envelope (NE) contribute to nucleocytoskeletal force transduction. A few NE proteins have been found to regulate the LINC complex. In this study, we identify one, Kuduk (Kud), which can reside at the outer nuclear membrane and is required for the development of Drosophila melanogaster ovarian follicles and NE morphology of myonuclei. Kud associates with LINC complex components in an evolutionarily conserved manner. Loss of Kud increases the level but impairs functioning of the LINC complex. Overexpression of Kud suppresses NE targeting of cytoskeleton-free LINC complexes. Thus, Kud acts as a quality control mechanism for LINC-mediated nucleocytoskeletal connections. Genetic data indicate that Kud also functions independently of the LINC complex. Overexpression of the human orthologue TMEM258 in Drosophila proved functional conservation. These findings expand our understanding of the regulation of LINC complexes and NE architecture.
© 2017 Ding et al.
Figures
Similar articles
-
LINCing complex functions at the nuclear envelope: what the molecular architecture of the LINC complex can reveal about its function.Nucleus. 2013 Jan-Feb;4(1):29-36. doi: 10.4161/nucl.23387. Epub 2013 Jan 1. Nucleus. 2013. PMID: 23324460 Free PMC article.
-
Integrity of the Linker of Nucleoskeleton and Cytoskeleton Is Required for Efficient Herpesvirus Nuclear Egress.J Virol. 2017 Sep 12;91(19):e00330-17. doi: 10.1128/JVI.00330-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28724767 Free PMC article.
-
Recruitment of BAF to the nuclear envelope couples the LINC complex to endoreplication.Development. 2020 Dec 13;147(23):dev191304. doi: 10.1242/dev.191304. Development. 2020. PMID: 33168584 Free PMC article.
-
LINC complex proteins in development and disease.Curr Top Dev Biol. 2014;109:287-321. doi: 10.1016/B978-0-12-397920-9.00004-4. Curr Top Dev Biol. 2014. PMID: 24947240 Review.
-
Life outside the LINC complex - Do SUN proteins have LINC-independent functions?Bioessays. 2024 Aug;46(8):e2400034. doi: 10.1002/bies.202400034. Epub 2024 May 27. Bioessays. 2024. PMID: 38798157 Review.
Cited by
-
tmem-258 is dispensable for both nuclear anchorage and migration in C. elegans.MicroPubl Biol. 2020 Jan 2;2020:10.17912/micropub.biology.000208. doi: 10.17912/micropub.biology.000208. MicroPubl Biol. 2020. PMID: 32550492 Free PMC article. No abstract available.
-
Purriato is a conserved small open reading frame gene that interacts with the CASA pathway to regulate muscle homeostasis and epithelial tissue growth in Drosophila.Front Cell Dev Biol. 2023 Mar 10;11:1117454. doi: 10.3389/fcell.2023.1117454. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36968202 Free PMC article.
-
Chain reaction: LINC complexes and nuclear positioning.F1000Res. 2019 Jan 31;8:F1000 Faculty Rev-136. doi: 10.12688/f1000research.16877.1. eCollection 2019. F1000Res. 2019. PMID: 30774932 Free PMC article. Review.
-
Nucleus Mechanosensing in Cardiomyocytes.Int J Mol Sci. 2023 Aug 28;24(17):13341. doi: 10.3390/ijms241713341. Int J Mol Sci. 2023. PMID: 37686151 Free PMC article. Review.
-
SUN/KASH interactions facilitate force transmission across the nuclear envelope.Nucleus. 2019 Dec;10(1):73-80. doi: 10.1080/19491034.2019.1595313. Nucleus. 2019. PMID: 30888237 Free PMC article. Review.
References
-
- Attali, R., Warwar N., Israel A., Gurt I., McNally E., Puckelwartz M., Glick B., Nevo Y., Ben-Neriah Z., and Melki J.. 2009. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Hum. Mol. Genet. 18:3462–3469. 10.1093/hmg/ddp290 - DOI - PubMed
-
- Bochenek, G., Häsler R., El Mokhtari N.E., König I.R., Loos B.G., Jepsen S., Rosenstiel P., Schreiber S., and Schaefer A.S.. 2013. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 22:4516–4527. 10.1093/hmg/ddt299 - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

