OsMPH1 regulates plant height and improves grain yield in rice
- PMID: 28708834
- PMCID: PMC5510837
- DOI: 10.1371/journal.pone.0180825
OsMPH1 regulates plant height and improves grain yield in rice
Abstract
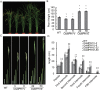
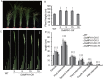
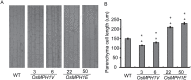
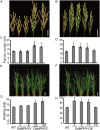
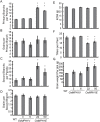
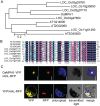
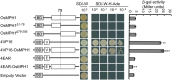
Plant height is a major trait affecting yield potential in rice. Using a large-scale hybrid transcription factor approach, we identified the novel MYB-like transcription factor OsMPH1 (MYB-like gene of Plant Height 1), which is involved in the regulation of plant height in rice. Overexpression of OsMPH1 leads to increases of plant height and grain yield in rice, while knockdown of OsMPH1 leads to the opposite phenotypes. Microscopy of longitudinal stem sections indicated that a change in internode cell length resulted in the change in plant height. RNA sequencing (RNA-seq) analysis of transgenic rice lines showed that multiple genes related to cell elongation and cell wall synthesis, which are associated with plant height and yield phenotypes, exhibited an altered expression profile. These results imply that OsMPH1 might be involved in specific recognition and signal transduction processes related to plant height and yield formation, providing further insights into the mechanisms underlying the regulation of plant height and providing a candidate gene for the efficient improvement of rice yield.
Conflict of interest statement
Figures


Similar articles
-
Regulation of plant height in rice by the Polycomb group genes OsEMF2b, OsFIE2 and OsCLF.Plant Sci. 2018 Feb;267:157-167. doi: 10.1016/j.plantsci.2017.11.007. Epub 2017 Nov 21. Plant Sci. 2018. PMID: 29362094
-
OsGASR9 positively regulates grain size and yield in rice (Oryza sativa).Plant Sci. 2019 Sep;286:17-27. doi: 10.1016/j.plantsci.2019.03.008. Epub 2019 Mar 26. Plant Sci. 2019. PMID: 31300138
-
Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice.Plant Physiol. 2013 Jan;161(1):317-29. doi: 10.1104/pp.112.208496. Epub 2012 Nov 2. Plant Physiol. 2013. PMID: 23124325 Free PMC article.
-
Two highly similar DEAD box proteins, OsRH2 and OsRH34, homologous to eukaryotic initiation factor 4AIII, play roles of the exon junction complex in regulating growth and development in rice.BMC Plant Biol. 2016 Apr 12;16:84. doi: 10.1186/s12870-016-0769-5. BMC Plant Biol. 2016. PMID: 27071313 Free PMC article.
-
New clues into the mechanisms of rice domestication.J Biosci. 2019 Jun;44(2):28. J Biosci. 2019. PMID: 31180041 Review.
Cited by
-
Prospects for the accelerated improvement of the resilient crop quinoa.J Exp Bot. 2020 Sep 19;71(18):5333-5347. doi: 10.1093/jxb/eraa285. J Exp Bot. 2020. PMID: 32643753 Free PMC article.
-
Detection of QTLs for Plant Height Architecture Traits in Rice (Oryza sativa L.) by Association Mapping and the RSTEP-LRT Method.Plants (Basel). 2022 Apr 6;11(7):999. doi: 10.3390/plants11070999. Plants (Basel). 2022. PMID: 35406978 Free PMC article.
-
Exploring the Relationships Between Yield and Yield-Related Traits for Rice Varieties Released in China From 1978 to 2017.Front Plant Sci. 2019 May 7;10:543. doi: 10.3389/fpls.2019.00543. eCollection 2019. Front Plant Sci. 2019. PMID: 31134107 Free PMC article.
-
An APETALA2/ethylene responsive factor transcription factor GmCRF4a regulates plant height and auxin biosynthesis in soybean.Front Plant Sci. 2022 Sep 6;13:983650. doi: 10.3389/fpls.2022.983650. eCollection 2022. Front Plant Sci. 2022. PMID: 36147224 Free PMC article.
-
Quantitative Trait Loci Mapping Identified Candidate Genes Involved in Plant Height Regulation in Rice.Int J Mol Sci. 2023 Nov 29;24(23):16895. doi: 10.3390/ijms242316895. Int J Mol Sci. 2023. PMID: 38069217 Free PMC article.
References
-
- Palme K, Li X, Teale WD (2014) Towards second green revolution: engineering nitrogen use efficiency. J Genet Genomics 41: 315–316. doi: 10.1016/j.jgg.2014.05.003 - DOI - PubMed
-
- Mooney BP (2009) The second green revolution? Production of plant-based biodegradable plastics. Biochem J 418: 219–232. doi: 10.1042/BJ20081769 - DOI - PubMed
-
- Wollenweber B, Porter JR, Lubberstedt T (2005) Need for multidisciplinary research towards a second green revolution. Curr Opin Plant Biol 8: 337–341. doi: 10.1016/j.pbi.2005.03.001 - DOI - PubMed
-
- Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, et al. (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13: 544 doi: 10.1186/1471-2164-13-544 - DOI - PMC - PubMed
-
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, et al. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581. doi: 10.1016/j.tplants.2010.06.005 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

