The function of DNA binding protein nucleophosmin in AAV replication
- PMID: 28704696
- PMCID: PMC5572047
- DOI: 10.1016/j.virol.2017.07.007
The function of DNA binding protein nucleophosmin in AAV replication
Abstract
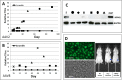
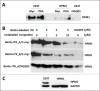
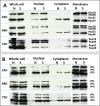
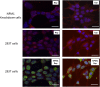
Adeno-associated viruses (AAV) contain minimal viral proteins necessary for their replication. During virus assembly, AAV acquire, inherently and submissively, various cellular proteins. Our previous studies identified the association of AAV vectors with the DNA binding protein nucleophosmin (NPM1). Nucleophosmin has been reported to enhance AAV infection by mobilizing AAV capsids into and out of the nucleolus, indicating the importance of NPM1 in the AAV life cycle; however the role of NPM1 in AAV production remains unknown. In this study, we systematically investigated NPM1 function on AAV production using NPM1 knockdown cells and revealing for the first time the presence of G-quadruplex DNA sequences (GQRS) in the AAV genome, the synergistic NPM1-GQRS function in AAV production and the significant enhancement of NPM1 gene knockdown on AAV vector production. Understanding the role of cellular proteins in the AAV life cycle will greatly facilitate high titre production of AAV vectors for clinical use.
Keywords: AAV biology; G-quadruplex DNA sequences; NPM1; shRNA.
Crown Copyright © 2017. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Establishment of a novel cell line for the enhanced production of recombinant adeno-associated virus vectors for gene therapy.Hum Gene Ther. 2014 Nov;25(11):929-41. doi: 10.1089/hum.2014.041. Epub 2014 Sep 11. Hum Gene Ther. 2014. PMID: 25072415 Free PMC article.
-
Identification of cellular proteins that interact with the adeno-associated virus rep protein.J Virol. 2009 Jan;83(1):454-69. doi: 10.1128/JVI.01939-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971280 Free PMC article.
-
Serotype-specific replicating AAV helper constructs increase recombinant AAV type 2 vector production.Virology. 2005 Apr 25;335(1):10-21. doi: 10.1016/j.virol.2005.02.008. Virology. 2005. PMID: 15823602
-
Implication of B23/NPM1 in Viral Infections, Potential Uses of B23/NPM1 Inhibitors as Antiviral Therapy.Infect Disord Drug Targets. 2019;19(1):2-16. doi: 10.2174/1871526518666180327124412. Infect Disord Drug Targets. 2019. PMID: 29589547 Review.
-
[Capsid assembly and DNA encapsidation of adeno-associated virus].Sheng Wu Gong Cheng Xue Bao. 2011 Apr;27(4):531-8. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 21847986 Review. Chinese.
Cited by
-
No G-Quadruplex Structures in the DNA of Parvovirus B19: Experimental Evidence versus Bioinformatic Predictions.Viruses. 2020 Aug 25;12(9):935. doi: 10.3390/v12090935. Viruses. 2020. PMID: 32854437 Free PMC article.
-
G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy.Nucleic Acids Res. 2018 Apr 20;46(7):3270-3283. doi: 10.1093/nar/gky187. Nucleic Acids Res. 2018. PMID: 29554280 Free PMC article. Review.
-
G-Quadruplexes in the Viral Genome: Unlocking Targets for Therapeutic Interventions and Antiviral Strategies.Viruses. 2023 Nov 5;15(11):2216. doi: 10.3390/v15112216. Viruses. 2023. PMID: 38005893 Free PMC article. Review.
-
Adeno-associated virus vector system controlling capsid expression improves viral quantity and quality.iScience. 2023 Mar 23;26(4):106487. doi: 10.1016/j.isci.2023.106487. eCollection 2023 Apr 21. iScience. 2023. PMID: 37096037 Free PMC article.
-
G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide.PLoS Comput Biol. 2018 Dec 13;14(12):e1006675. doi: 10.1371/journal.pcbi.1006675. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30543627 Free PMC article.
References
-
- Banuelos S., Lectez B., Taneva S.G., Ormaza G., Alonso-Marino M., Calle X., Urbaneja M.A. Recognition of intermolecular G-quadruplexes by full length nucleophosmin. Effect of a leukaemia-associated mutation. FEBS Lett. 2013;587(14):2254–2259. - PubMed
-
- Brument N., Morenweiser R., Blouin V., Toublanc E., Raimbaud I., Cherel Y., Folliot S., Gaden F., Boulanger P., Kroner-Lux G., Moullier P., Rolling F., Salvetti A. A versatile and scalable two-step ion-exchange chromatography process for the purification of recombinant adeno-associated virus serotypes-2 and −5. Mol. Ther.: J. Am. Soc. Gene Ther. 2002;6(5):678–686. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

