Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication
- PMID: 28693575
- PMCID: PMC5504599
- DOI: 10.1186/s12985-017-0794-5
Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication
Abstract
Background: Porcine reproductive and respiratory syndrome virus (PRRSV) causes reproductive failures in sows and respiratory diseases in growing pigs, resulting in huge economic loss for the pig production worldwide. The nonstructural protein 9 (nsp9) and nonstructural protein 2 (nsp2) of PRRSV are known to play important roles in viral replication. Cellular interleukin-2 enhancer binding factor 2 (ILF2) participates in many cellular pathways and involves in life cycle of some viruses. In the present study, we analyzed the interaction of cellular ILF2 with the nsp9 and nsp2 of PRRSV in vitro and explored the effect of ILF2 on viral replication.
Methods: The interaction of ILF2 with the nsp9 or nsp2 of PRRSV was analyzed in 293FT cells and MARC-145 cells by co-immunoprecipitation (Co-IP) and the co-localization of ILF2 with the nsp9 or nsp2 of PRRSV in MARC-145 cell and pulmonary alveolar macrophages (PAMs) was examined by confocal immunofluorescence assay. The effect of ILF2 knockdown and over-expression on PRRSV replication was explored in MARC-145 cells by small interfering RNA (siRNA) and lentivirus transduction, respectively.
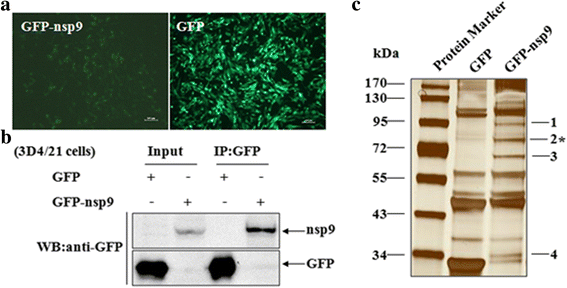
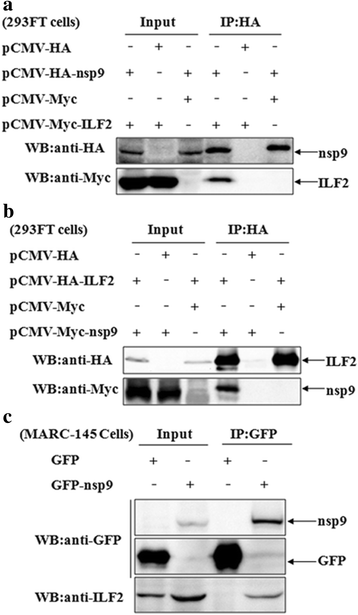
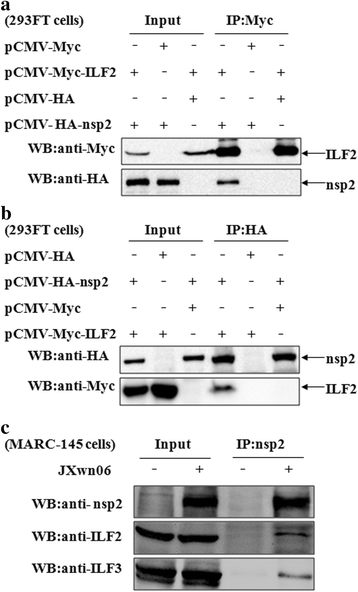
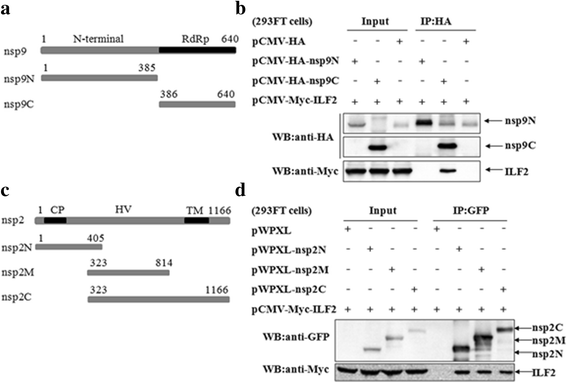
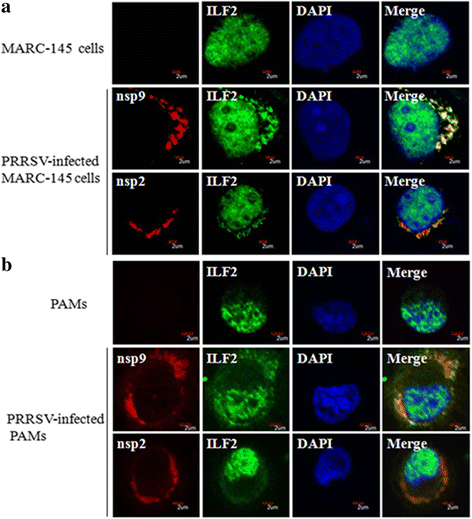
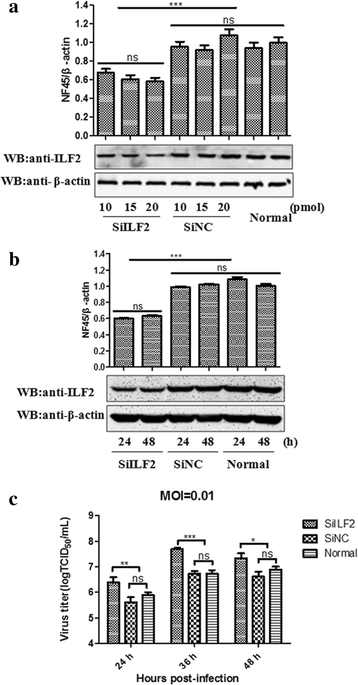
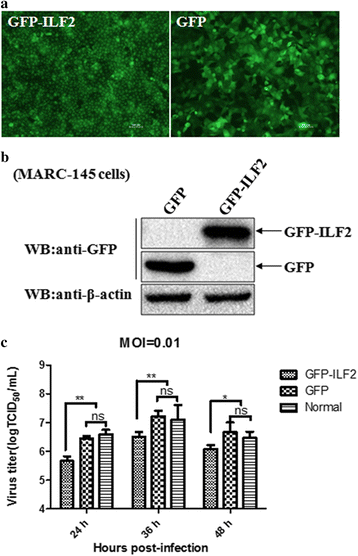
Results: The interaction of ILF2 with nsp9 or nsp2 was first demonstrated in 293FT cells co-transfected with ILF2-expressing plasmid and nsp9-expressing plasmid or nsp2-expressing plasmid. The interaction of endogenous ILF2 with the nsp9 or nsp2 of PRRSV was further confirmed in MARC-145 cells transduced with GFP-nsp9-expressing lentiviruses or infected with PRRSV JXwn06. The RdRp domain of nsp9 was shown to be responsible for its interaction with ILF2, while three truncated nsp2 were shown to interact with ILF2. Moreover, we observed that ILF2 partly translocated from the nucleus to the cytoplasm and co-localized with nsp9 and nsp2 in PRRSV-infected MARC-145 cells and PAMs. Finally, our analysis indicated that knockdown of ILF2 favored the replication of PRRSV, while over-expression of ILF2 impaired the viral replication in MARC-145 cells.
Conclusion: Our findings are the first to confirm that the porcine ILF2 interacts with the nsp9 and nsp2 of PRRSV in vitro, and exerts negatively regulatory effect on the replication of PRRSV. Our present study provides more evidence for understanding the roles of the interactions between cellular proteins and viral proteins in the replication of PRRSV.
Keywords: Interaction; Interleukin-2 enhancer binding factor 2 (ILF2); Nonstructural protein 2 (nsp2); Nonstructural protein 9 (nsp9); Porcine reproductive and respiratory syndrome virus (PRRSV); Replication.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Figures
Similar articles
-
The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro.Virus Res. 2015 Jan 2;195:217-24. doi: 10.1016/j.virusres.2014.10.021. Epub 2014 Nov 1. Virus Res. 2015. PMID: 25449571 Free PMC article.
-
The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus.Virus Res. 2014 Aug 30;189:106-13. doi: 10.1016/j.virusres.2014.05.015. Epub 2014 May 26. Virus Res. 2014. PMID: 24874199
-
The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro.Virology. 2014 Sep;464-465:432-440. doi: 10.1016/j.virol.2014.07.036. Epub 2014 Aug 21. Virology. 2014. PMID: 25146601 Free PMC article.
-
Non-structural protein 2 of the porcine reproductive and respiratory syndrome (PRRS) virus: a crucial protein in viral pathogenesis, immunity and diagnosis.Res Vet Sci. 2013 Aug;95(1):1-7. doi: 10.1016/j.rvsc.2013.03.015. Epub 2013 Apr 13. Res Vet Sci. 2013. PMID: 23591056 Review.
-
The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins.Virus Res. 2010 Dec;154(1-2):61-76. doi: 10.1016/j.virusres.2010.07.030. Epub 2010 Aug 7. Virus Res. 2010. PMID: 20696193 Free PMC article. Review.
Cited by
-
Interleukin-2 enhancer binding factor 2 negatively regulates the replication of duck hepatitis A virus type 1 by disrupting the RNA-dependent RNA polymerase activity of 3D polymerase.Vet Res. 2024 Mar 26;55(1):40. doi: 10.1186/s13567-024-01294-x. Vet Res. 2024. PMID: 38532469 Free PMC article.
-
TRIM4-mediated ubiquitination of NSP2 restricts porcine reproductive and respiratory syndrome virus proliferation.BMC Vet Res. 2022 May 30;18(1):208. doi: 10.1186/s12917-022-03309-1. BMC Vet Res. 2022. PMID: 35637527 Free PMC article.
-
Research Progress on the NSP9 Protein of Porcine Reproductive and Respiratory Syndrome Virus.Front Vet Sci. 2022 Jul 11;9:872205. doi: 10.3389/fvets.2022.872205. eCollection 2022. Front Vet Sci. 2022. PMID: 35898550 Free PMC article. Review.
-
Mapping the Nonstructural Protein Interaction Network of Porcine Reproductive and Respiratory Syndrome Virus.J Virol. 2018 Nov 27;92(24):e01112-18. doi: 10.1128/JVI.01112-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30282705 Free PMC article.
-
Enterovirus 71 Represses Interleukin Enhancer-Binding Factor 2 Production and Nucleus Translocation to Antagonize ILF2 Antiviral Effects.Viruses. 2019 Dec 23;12(1):22. doi: 10.3390/v12010022. Viruses. 2019. PMID: 31878072 Free PMC article.
References
-
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

