Cooperation between Strain-Specific and Broadly Neutralizing Responses Limited Viral Escape and Prolonged the Exposure of the Broadly Neutralizing Epitope
- PMID: 28679760
- PMCID: PMC5571269
- DOI: 10.1128/JVI.00828-17
Cooperation between Strain-Specific and Broadly Neutralizing Responses Limited Viral Escape and Prolonged the Exposure of the Broadly Neutralizing Epitope
Erratum in
-
Correction for Anthony et al., "Cooperation between Strain-Specific and Broadly Neutralizing Responses Limited Viral Escape and Prolonged the Exposure of the Broadly Neutralizing Epitope".J Virol. 2018 Apr 13;92(9):e00243-18. doi: 10.1128/JVI.00243-18. Print 2018 May 1. J Virol. 2018. PMID: 29654221 Free PMC article. No abstract available.
Abstract
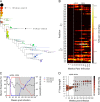
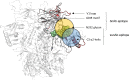
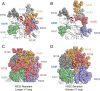
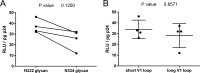
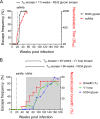
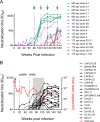
V3-glycan-targeting broadly neutralizing antibodies (bNAbs) are a focus of HIV-1 vaccine development. Understanding the viral dynamics that stimulate the development of these antibodies can provide insights for immunogen design. We used a deep-sequencing approach, together with neutralization phenotyping, to investigate the rate and complexity of escape from V3-glycan-directed bNAbs compared to overlapping early strain-specific neutralizing antibody (ssNAb) responses to the V3/C3 region in donor CAP177. Escape from the ssNAb response occurred rapidly via an N334-to-N332 glycan switch, which took just 7.5 weeks to reach >50% frequency. In contrast, escape from the bNAbs was mediated via multiple pathways and took longer, with escape first occurring through an increase in V1 loop length, which took 46 weeks to reach 50% frequency, followed by an N332-to-N334 reversion, which took 66 weeks. Importantly, bNAb escape was incomplete, with contemporaneous neutralization observed up to 3 years postinfection. Both the ssNAb response and the bNAb response were modulated by the presence/absence of the N332 glycan, indicating an overlap between the two epitopes. Thus, selective pressure by ssNAbs to maintain the N332 glycan may have constrained the bNAb escape pathway. This slower and incomplete viral escape resulted in prolonged exposure of the bNAb epitope, which may in turn have aided the maturation of the bNAb lineage.IMPORTANCE The development of an HIV-1 vaccine is of paramount importance, and broadly neutralizing antibodies are likely to be a key component of a protective vaccine. The V3-glycan-targeting bNAb responses are among the most promising vaccine targets, as they are commonly elicited during infection. Understanding the interplay between viral evolution and the development of these antibodies provides insights that may guide immunogen design. Our work contrasted the dynamics of the early strain-specific antibodies and the later broadly neutralizing responses to a common Env target (V3C3), showing slower and more complex escape from bNAbs. Constrained bNAb escape, together with evidence of contemporaneous autologous virus neutralization, supports the proposal that prolonged exposure of the bNAb epitope enabled the maturation of the bNAb lineage.
Keywords: N332 glycan; V3-glycan supersite; broadly neutralizing antibodies; deep sequencing; glycan holes; glycan shield; helper/cooperating NAb responses; neutralization escape; viral escape.
Copyright © 2017 Anthony et al.
Figures

Similar articles
-
A Rare Mutation in an Infant-Derived HIV-1 Envelope Glycoprotein Alters Interprotomer Stability and Susceptibility to Broadly Neutralizing Antibodies Targeting the Trimer Apex.J Virol. 2020 Sep 15;94(19):e00814-20. doi: 10.1128/JVI.00814-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669335 Free PMC article.
-
Emergence of broadly neutralizing antibodies and viral coevolution in two subjects during the early stages of infection with human immunodeficiency virus type 1.J Virol. 2014 Nov;88(22):12968-81. doi: 10.1128/JVI.01816-14. Epub 2014 Aug 13. J Virol. 2014. PMID: 25122781 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
Targeting the N332-supersite of the HIV-1 envelope for vaccine design.Expert Opin Ther Targets. 2020 Jun;24(6):499-509. doi: 10.1080/14728222.2020.1752183. Epub 2020 Apr 27. Expert Opin Ther Targets. 2020. PMID: 32340497 Free PMC article. Review.
-
Strategies for induction of HIV-1 envelope-reactive broadly neutralizing antibodies.J Int AIDS Soc. 2021 Nov;24 Suppl 7(Suppl 7):e25831. doi: 10.1002/jia2.25831. J Int AIDS Soc. 2021. PMID: 34806332 Free PMC article. Review.
Cited by
-
Replication in the presence of dengue convalescent serum impacts Zika virus neutralization sensitivity and fitness.Front Cell Infect Microbiol. 2023 Mar 9;13:1130749. doi: 10.3389/fcimb.2023.1130749. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36968111 Free PMC article.
-
Selective transmission of some HIV-1 subtype C variants might depend on Envelope stimulating dendritic cells to secrete IL-10.PLoS One. 2020 Jan 24;15(1):e0227533. doi: 10.1371/journal.pone.0227533. eCollection 2020. PLoS One. 2020. PMID: 31978062 Free PMC article.
-
Prevalence of resistance-associated viral variants to the HIV-specific broadly neutralising antibody 10-1074 in a UK bNAb-naïve population.Front Immunol. 2024 Mar 18;15:1352123. doi: 10.3389/fimmu.2024.1352123. eCollection 2024. Front Immunol. 2024. PMID: 38562938 Free PMC article.
-
The expanding array of HIV broadly neutralizing antibodies.Retrovirology. 2018 Oct 16;15(1):70. doi: 10.1186/s12977-018-0453-y. Retrovirology. 2018. PMID: 30326938 Free PMC article. Review.
-
Neutralization Breadth and Potency of Single-Chain Variable Fragments Derived from Broadly Neutralizing Antibodies Targeting Multiple Epitopes on the HIV-1 Envelope.J Virol. 2020 Jan 6;94(2):e01533-19. doi: 10.1128/JVI.01533-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31619559 Free PMC article.
References
-
- Liao H- X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin JC, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi:10.1038/nature12053. - DOI - PMC - PubMed
-
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi:10.1038/nature13036. - DOI - PMC - PubMed
-
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang KK, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao HX, Kwong PD, Haynes BF. 2016. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165:449–463. doi:10.1016/j.cell.2016.02.022. - DOI - PMC - PubMed
-
- MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang C-H, Ramos A, Bian CB, Wickramasinghe L, Kong L, Eren K, Wu C-Y, Wong C-H, IAVI Protocol C Investigators & The IAVI African HIV Research Network, Kosakovsky Pond SL, Wilson IA, Burton DR, Poignard P. 2016. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 44:1215–1226. doi:10.1016/j.immuni.2016.04.016. - DOI - PMC - PubMed
-
- Garces F, Lee JH, de Val N, Torrents de la Pena A, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43:1053–1063. doi:10.1016/j.immuni.2015.11.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

