Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues
- PMID: 28662667
- PMCID: PMC5490202
- DOI: 10.1186/s12915-017-0397-z
Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues
Abstract
Background: Strigolactones (SLs) are a class of plant hormones that control many aspects of plant growth. The SL signalling mechanism is homologous to that of karrikins (KARs), smoke-derived compounds that stimulate seed germination. In angiosperms, the SL receptor is an α/β-hydrolase known as DWARF14 (D14); its close homologue, KARRIKIN INSENSITIVE2 (KAI2), functions as a KAR receptor and likely recognizes an uncharacterized, endogenous signal ('KL'). Previous phylogenetic analyses have suggested that the KAI2 lineage is ancestral in land plants, and that canonical D14-type SL receptors only arose in seed plants; this is paradoxical, however, as non-vascular plants synthesize and respond to SLs.
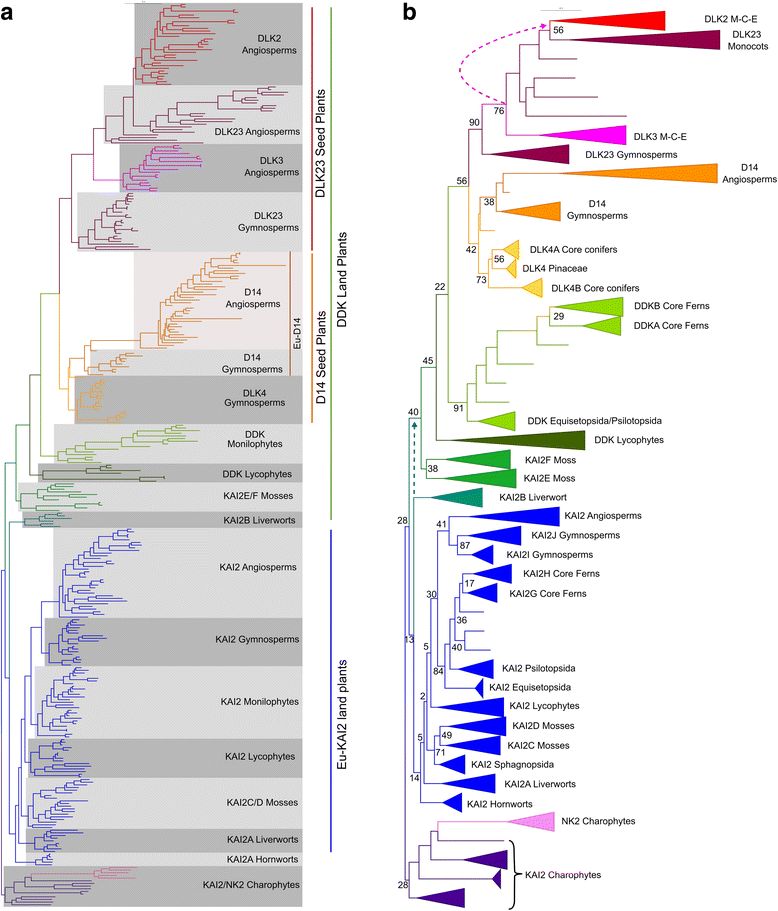
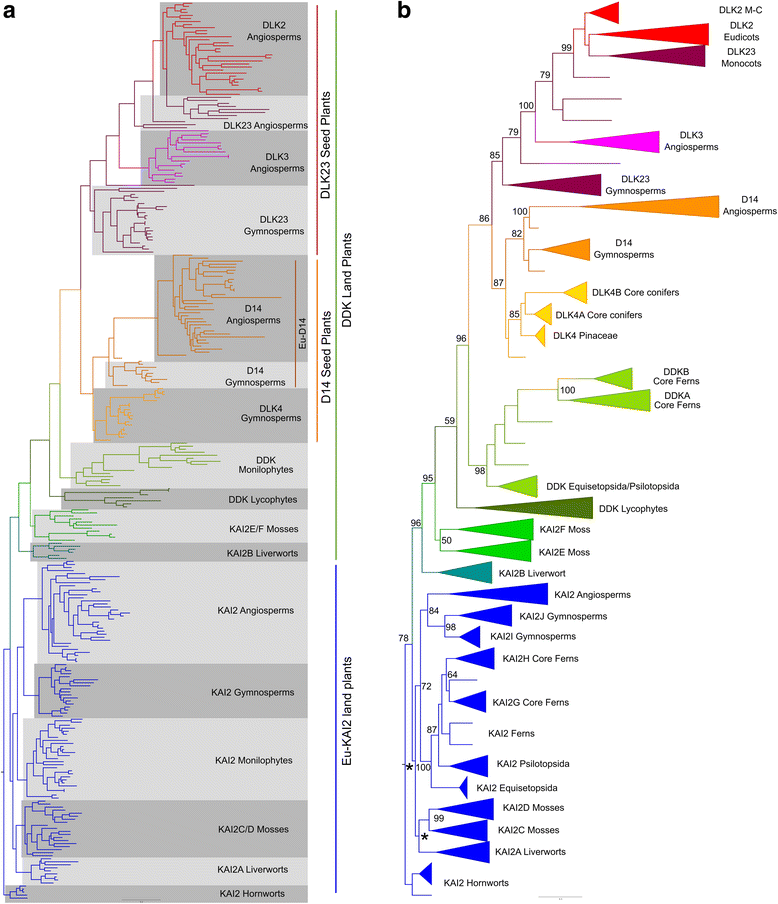
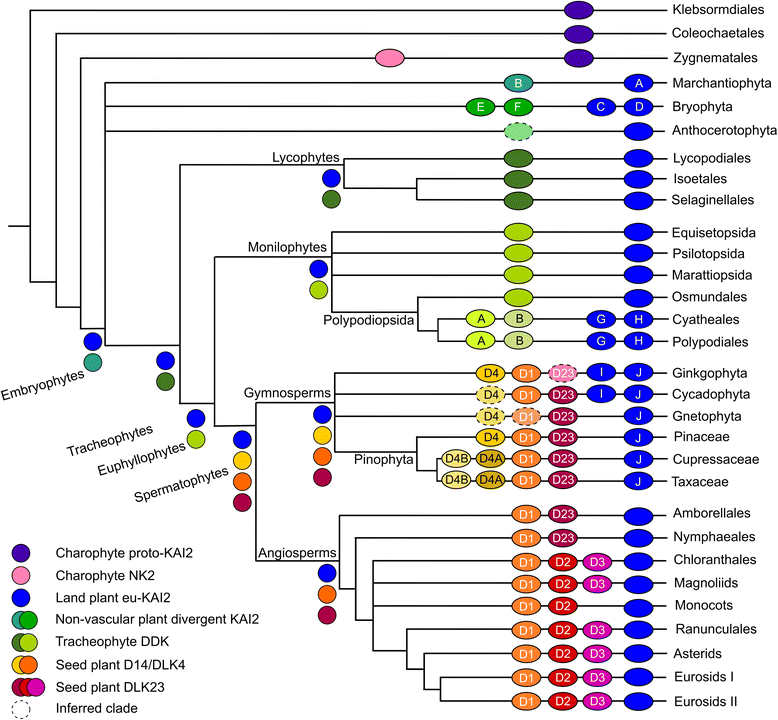
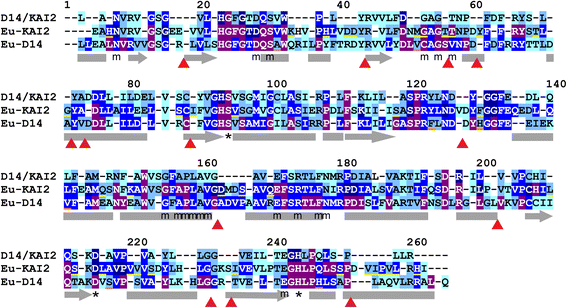
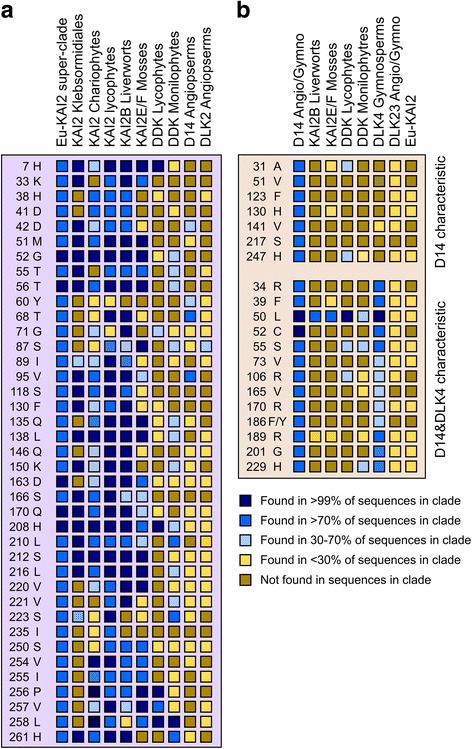
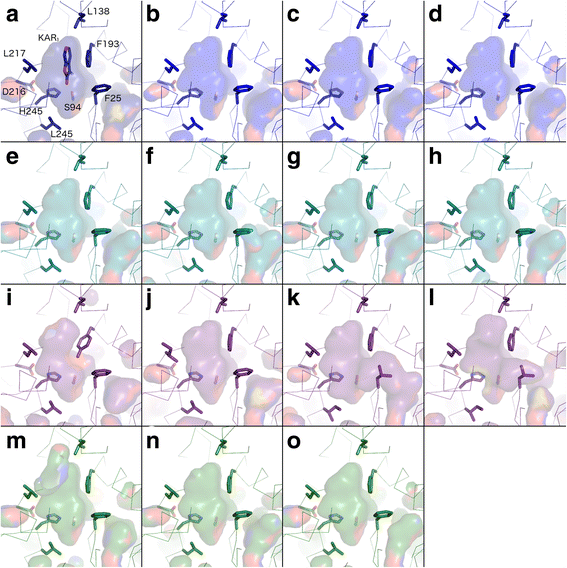
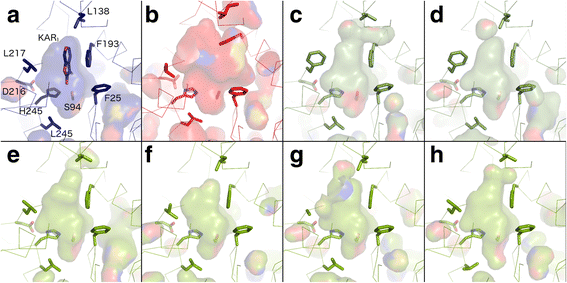
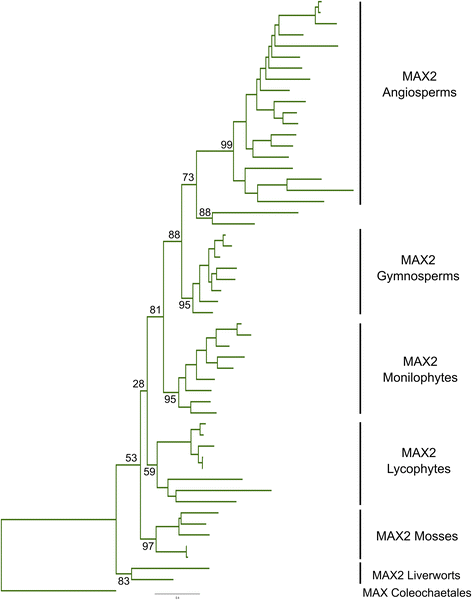
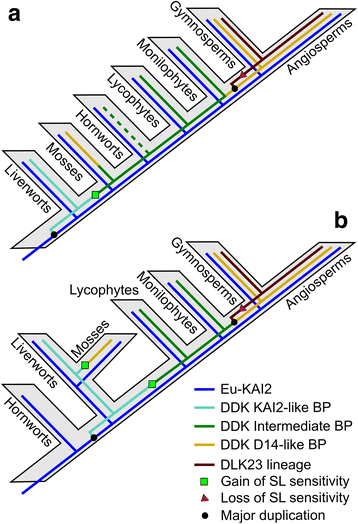
Results: We have used a combination of phylogenetic and structural approaches to re-assess the evolution of the D14/KAI2 family in land plants. We analysed 339 members of the D14/KAI2 family from land plants and charophyte algae. Our phylogenetic analyses show that the divergence between the eu-KAI2 lineage and the DDK (D14/DLK2/KAI2) lineage that includes D14 occurred very early in land plant evolution. We show that eu-KAI2 proteins are highly conserved, and have unique features not found in DDK proteins. Conversely, we show that DDK proteins show considerable sequence and structural variation to each other, and lack clearly definable characteristics. We use homology modelling to show that the earliest members of the DDK lineage structurally resemble KAI2 and that SL receptors in non-seed plants likely do not have D14-like structure. We also show that certain groups of DDK proteins lack the otherwise conserved MORE AXILLARY GROWTH2 (MAX2) interface, and may thus function independently of MAX2, which we show is highly conserved throughout land plant evolution.
Conclusions: Our results suggest that D14-like structure is not required for SL perception, and that SL perception has relatively relaxed structural requirements compared to KAI2-mediated signalling. We suggest that SL perception gradually evolved by neo-functionalization within the DDK lineage, and that the transition from KAI2-like to D14-like protein may have been driven by interactions with protein partners, rather than being required for SL perception per se.
Keywords: Neo-functionalization; Phylogenetics; Strigolactone evolution; Strigolactone signalling.
Figures
Similar articles
-
Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens.Planta. 2016 Jun;243(6):1441-53. doi: 10.1007/s00425-016-2481-y. Epub 2016 Mar 15. Planta. 2016. PMID: 26979323
-
Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may Perceive an Unknown Signal that is not Karrikin or Strigolactone.Front Plant Sci. 2016 Jan 8;6:1219. doi: 10.3389/fpls.2015.01219. eCollection 2015. Front Plant Sci. 2016. PMID: 26779242 Free PMC article.
-
Molecular evolution and diversification of the SMXL gene family.J Exp Bot. 2018 Apr 23;69(9):2367-2378. doi: 10.1093/jxb/ery097. J Exp Bot. 2018. PMID: 29538714
-
The origins and mechanisms of karrikin signalling.Curr Opin Plant Biol. 2013 Oct;16(5):667-73. doi: 10.1016/j.pbi.2013.07.005. Epub 2013 Aug 14. Curr Opin Plant Biol. 2013. PMID: 23954000 Review.
-
Origins of strigolactone and karrikin signaling in plants.Trends Plant Sci. 2022 May;27(5):450-459. doi: 10.1016/j.tplants.2021.11.009. Epub 2021 Dec 4. Trends Plant Sci. 2022. PMID: 34876337 Review.
Cited by
-
KAI2 regulates seedling development by mediating light-induced remodelling of auxin transport.New Phytol. 2022 Jul;235(1):126-140. doi: 10.1111/nph.18110. Epub 2022 Apr 9. New Phytol. 2022. PMID: 35313031 Free PMC article.
-
The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice.Nat Commun. 2020 Apr 30;11(1):2114. doi: 10.1038/s41467-020-16021-1. Nat Commun. 2020. PMID: 32355217 Free PMC article.
-
Rice DWARF14 acts as an unconventional hormone receptor for strigolactone.J Exp Bot. 2018 Apr 23;69(9):2355-2365. doi: 10.1093/jxb/ery014. J Exp Bot. 2018. PMID: 29365172 Free PMC article.
-
The mechanism of host-induced germination in root parasitic plants.Plant Physiol. 2021 Apr 23;185(4):1353-1373. doi: 10.1093/plphys/kiab043. Plant Physiol. 2021. PMID: 33793958 Free PMC article. Review.
-
SUPPRESSOR OF MAX2 1-LIKE (SMXL) homologs are MAX2-dependent repressors of Physcomitrium patens growth.Plant Cell. 2024 May 1;36(5):1655-1672. doi: 10.1093/plcell/koae009. Plant Cell. 2024. PMID: 38242840 Free PMC article.
References
-
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, et al. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155:974–87. doi: 10.1104/pp.110.164640. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

