Diversity and evolution of mariner-like elements in aphid genomes
- PMID: 28662628
- PMCID: PMC5490172
- DOI: 10.1186/s12864-017-3856-6
Diversity and evolution of mariner-like elements in aphid genomes
Abstract
Background: Although transposons have been identified in almost all organisms, genome-wide information on mariner elements in Aphididae remains unknown. Genomes of Acyrthosiphon pisum, Diuraphis noxia and Myzus persicae belonging to the Macrosiphini tribe, actually available in databases, have been investigated.
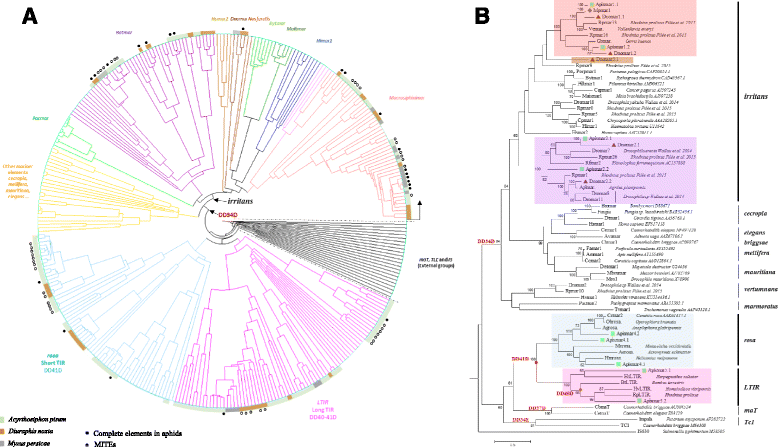
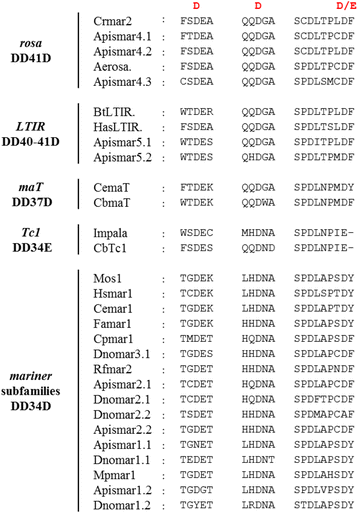
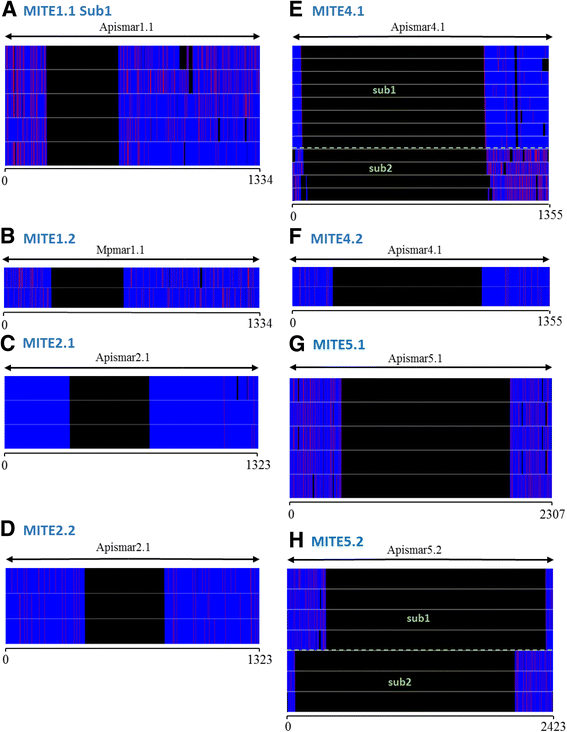
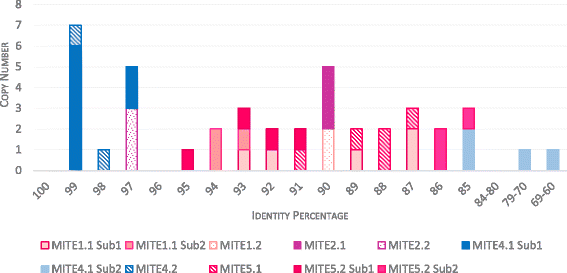
Results: A total of 22 lineages were identified. Classification and phylogenetic analysis indicated that they were subdivided into three monophyletic groups, each of them containing at least one putative complete sequence, and several non-autonomous sublineages corresponding to Miniature Inverted-Repeat Transposable Elements (MITE), probably generated by internal deletions. A high proportion of truncated and dead copies was also detected. The three clusters can be defined from their catalytic site: (i) mariner DD34D, including three subgroups of the irritans subfamily (Macrosiphinimar, Batmar-like elements and Dnomar-like elements); (ii) rosa DD41D, found in A. pisum and D. noxia; (iii) a new clade which differs from rosa through long TIRs and thus designated LTIR-like elements. Based on its catalytic domain, this new clade is subdivided into DD40D and DD41D subgroups. Compared to other Tc1/mariner superfamily sequences, rosa DD41D and LTIR DD40-41D seem more related to maT DD37D family.
Conclusion: Overall, our results reveal three clades belonging to the irritans subfamily, rosa and new LTIR-like elements. Data on structure and specific distribution of these transposable elements in the Macrosiphini tribe contribute to the understanding of their evolutionary history and to that of their hosts.
Keywords: Aphids; Comparative genomics; MITEs; Molecular evolution; Tc1-mariner; Transposable elements.
Figures

Similar articles
-
Divergent evolution profiles of DD37D and DD39D families of Tc1/mariner transposons in eukaryotes.Mol Phylogenet Evol. 2021 Aug;161:107143. doi: 10.1016/j.ympev.2021.107143. Epub 2021 Mar 10. Mol Phylogenet Evol. 2021. PMID: 33713798
-
Characterization of Mariner transposons in seven species of Rhus gall aphids.Sci Rep. 2021 Aug 11;11(1):16349. doi: 10.1038/s41598-021-95843-5. Sci Rep. 2021. PMID: 34381125 Free PMC article.
-
Genome-wide identification and evolution of TC1/Mariner in the silkworm (Bombyx mori) genome.Genes Genomics. 2018 May;40(5):485-495. doi: 10.1007/s13258-018-0648-6. Epub 2018 Feb 3. Genes Genomics. 2018. PMID: 29892960
-
Miniature inverted-repeat transposable elements: discovery, distribution, and activity.Genome. 2013 Sep;56(9):475-86. doi: 10.1139/gen-2012-0174. Epub 2013 Mar 8. Genome. 2013. PMID: 24168668 Review.
-
Modern thoughts on an ancyent marinere: function, evolution, regulation.Annu Rev Genet. 1997;31:337-58. doi: 10.1146/annurev.genet.31.1.337. Annu Rev Genet. 1997. PMID: 9442899 Review.
Cited by
-
Genome assembly of the Pendlebury's roundleaf bat, Hipposideros pendleburyi, revealed the expansion of Tc1/Mariner DNA transposons in Rhinolophoidea.DNA Res. 2022 Aug 23;29(5):dsac026. doi: 10.1093/dnares/dsac026. DNA Res. 2022. PMID: 36214371 Free PMC article.
-
Multiple Invasions of Visitor, a DD41D Family of Tc1/mariner Transposons, throughout the Evolution of Vertebrates.Genome Biol Evol. 2020 Jul 1;12(7):1060-1073. doi: 10.1093/gbe/evaa135. Genome Biol Evol. 2020. PMID: 32602886 Free PMC article.
-
Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects.BMC Evol Biol. 2019 Jan 9;19(1):11. doi: 10.1186/s12862-018-1324-9. BMC Evol Biol. 2019. PMID: 30626321 Free PMC article.
-
Mobilome of the Rhus Gall Aphid Schlechtendalia chinensis Provides Insight into TE Insertion-Related Inactivation of Functional Genes.Int J Mol Sci. 2022 Dec 15;23(24):15967. doi: 10.3390/ijms232415967. Int J Mol Sci. 2022. PMID: 36555609 Free PMC article.
-
Closing the Gap: Horizontal Transfer of Mariner Transposons between Rhus Gall Aphids and Other Insects.Biology (Basel). 2022 May 10;11(5):731. doi: 10.3390/biology11050731. Biology (Basel). 2022. PMID: 35625459 Free PMC article.
References
-
- Feschotte C, Zhang X, Wessler SR. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. In Mobile DNA II. Washington DC: ASM Press; 2002. pp. 1147–1158.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

