Translation Initiation from Conserved Non-AUG Codons Provides Additional Layers of Regulation and Coding Capacity
- PMID: 28655822
- PMCID: PMC5487733
- DOI: 10.1128/mBio.00844-17
Translation Initiation from Conserved Non-AUG Codons Provides Additional Layers of Regulation and Coding Capacity
Abstract
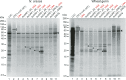
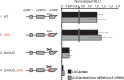
Neurospora crassa cpc-1 and Saccharomyces cerevisiae GCN4 are homologs specifying transcription activators that drive the transcriptional response to amino acid limitation. The cpc-1 mRNA contains two upstream open reading frames (uORFs) in its >700-nucleotide (nt) 5' leader, and its expression is controlled at the level of translation in response to amino acid starvation. We used N. crassa cell extracts and obtained data indicating that cpc-1 uORF1 and uORF2 are functionally analogous to GCN4 uORF1 and uORF4, respectively, in controlling translation. We also found that the 5' region upstream of the main coding sequence of the cpc-1 mRNA extends for more than 700 nucleotides without any in-frame stop codon. For 100 cpc-1 homologs from Pezizomycotina and from selected Basidiomycota, 5' conserved extensions of the CPC1 reading frame are also observed. Multiple non-AUG near-cognate codons (NCCs) in the CPC1 reading frame upstream of uORF2, some deeply conserved, could potentially initiate translation. At least four NCCs initiated translation in vitroIn vivo data were consistent with initiation at NCCs to produce N-terminally extended N. crassa CPC1 isoforms. The pivotal role played by CPC1, combined with its translational regulation by uORFs and NCC utilization, underscores the emerging significance of noncanonical initiation events in controlling gene expression.IMPORTANCE There is a deepening and widening appreciation of the diverse roles of translation in controlling gene expression. A central fungal transcription factor, the best-studied example of which is Saccharomyces cerevisiae GCN4, is crucial for the response to amino acid limitation. Two upstream open reading frames (uORFs) in the GCN4 mRNA are critical for controlling GCN4 synthesis. We observed that two uORFs in the corresponding Neurospora crassa cpc-1 mRNA appear functionally analogous to the GCN4 uORFs. We also discovered that, surprisingly, unlike GCN4, the CPC1 coding sequence extends far upstream from the presumed AUG start codon with no other in-frame AUG codons. Similar extensions were seen in homologs from many filamentous fungi. We observed that multiple non-AUG near-cognate codons (NCCs) in this extended reading frame, some conserved, initiated translation to produce longer forms of CPC1, underscoring the significance of noncanonical initiation in controlling gene expression.
Keywords: Neurospora; filamentous fungi; gene regulation; molecular genetics; translational control.
Copyright © 2017 Ivanov et al.
Figures



Comment in
-
Near-Cognate Codons Contribute Complexity to Translation Regulation.mBio. 2017 Nov 7;8(6):e01820-17. doi: 10.1128/mBio.01820-17. mBio. 2017. PMID: 29114030 Free PMC article.
Similar articles
-
Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control.Mol Cell Biol. 1991 Jan;11(1):486-96. doi: 10.1128/mcb.11.1.486-496.1991. Mol Cell Biol. 1991. PMID: 1986242 Free PMC article.
-
A quantitative model for translational control of the GCN4 gene of Saccharomyces cerevisiae.New Biol. 1991 May;3(5):511-24. New Biol. 1991. PMID: 1883814
-
Physical evidence for distinct mechanisms of translational control by upstream open reading frames.EMBO J. 2001 Nov 15;20(22):6453-63. doi: 10.1093/emboj/20.22.6453. EMBO J. 2001. PMID: 11707416 Free PMC article.
-
Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi.Annu Rev Microbiol. 2009;63:385-409. doi: 10.1146/annurev.micro.62.081307.162835. Annu Rev Microbiol. 2009. PMID: 19514854 Review.
-
Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2.Mol Microbiol. 1993 Oct;10(2):215-23. doi: 10.1111/j.1365-2958.1993.tb01947.x. Mol Microbiol. 1993. PMID: 7934812 Review.
Cited by
-
Non-AUG translation: a new start for protein synthesis in eukaryotes.Genes Dev. 2017 Sep 1;31(17):1717-1731. doi: 10.1101/gad.305250.117. Genes Dev. 2017. PMID: 28982758 Free PMC article. Review.
-
JUN mRNA Translation Regulation is Mediated by Multiple 5' UTR and Start Codon Features.bioRxiv [Preprint]. 2023 Nov 17:2023.11.17.567602. doi: 10.1101/2023.11.17.567602. bioRxiv. 2023. Update in: PLoS One. 2024 Mar 14;19(3):e0299779. doi: 10.1371/journal.pone.0299779. PMID: 38014201 Free PMC article. Updated. Preprint.
-
Understanding small ORF diversity through a comprehensive transcription feature classification.DNA Res. 2021 Sep 13;28(5):dsab007. doi: 10.1093/dnares/dsab007. DNA Res. 2021. PMID: 34240112 Free PMC article. Review.
-
Quantitative global studies reveal differential translational control by start codon context across the fungal kingdom.Nucleic Acids Res. 2020 Mar 18;48(5):2312-2331. doi: 10.1093/nar/gkaa060. Nucleic Acids Res. 2020. PMID: 32020195 Free PMC article.
-
Circadian clock control of eIF2α phosphorylation is necessary for rhythmic translation initiation.Proc Natl Acad Sci U S A. 2020 May 19;117(20):10935-10945. doi: 10.1073/pnas.1918459117. Epub 2020 Apr 30. Proc Natl Acad Sci U S A. 2020. PMID: 32355000 Free PMC article.
References
-
- Sachs MS. 1996. General and cross-pathway controls of amino acid biosynthesis, p 315–345. In Brambl R, Marzluf GA (ed), The Mycota: biochemistry and molecular biology, vol III Springer-Verlag, Heidelberg, Germany.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

