Development of Kras mutant lung adenocarcinoma in mice with knockout of the airway lineage-specific gene Gprc5a
- PMID: 28653505
- PMCID: PMC5774849
- DOI: 10.1002/ijc.30851
Development of Kras mutant lung adenocarcinoma in mice with knockout of the airway lineage-specific gene Gprc5a
Abstract
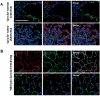
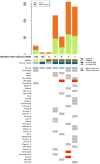
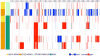
Despite the urgency for prevention and treatment of lung adenocarcinoma (LUAD), we still do not know drivers in pathogenesis of the disease. Earlier work revealed that mice with knockout of the G-protein coupled receptor Gprc5a develop late onset lung tumors including LUADs. Here, we sought to further probe the impact of Gprc5a expression on LUAD pathogenesis. We first surveyed GPRC5A expression in human tissues and found that GPRC5A was markedly elevated in human normal lung relative to other normal tissues and was consistently downregulated in LUADs. In sharp contrast to wild-type littermates, Gprc5a-/- mice treated chronically with the nicotine-specific carcinogen NNK developed LUADs by 6 months following NNK exposure. Immunofluorescence analysis revealed that the LUADs exhibited abundant expression of surfactant protein C and lacked the clara cell marker Ccsp, suggesting that these LUADs originated from alveolar type II cells. Next, we sought to survey genome-wide alterations in the pathogenesis of Gprc5a-/- LUADs. Using whole exome sequencing, we found that carcinogen-induced LUADs exhibited markedly higher somatic mutation burdens relative to spontaneous tumors. All LUADs were found to harbor somatic mutations in the Kras oncogene (p. G12D or p. Q61R). In contrast to spontaneous lesions, carcinogen-induced Gprc5a-/- LUADs exhibited mutations (variants and copy number gains) in additional drivers (Atm, Kmt2d, Nf1, Trp53, Met, Ezh2). Our study underscores genomic alterations that represent early events in the development of Kras mutant LUAD following Gprc5a loss and tobacco carcinogen exposure and that may constitute targets for prevention and early treatment of this disease.
Keywords: Gprc5a; Kras; carcinogenesis; lung adenocarcinoma; whole-exome sequencing.
© 2017 UICC.
Conflict of interest statement
The authors report no conflicts of interests.
Figures


Similar articles
-
Genome-Wide and Phenotypic Evaluation of Stem Cell Progenitors Derived From Gprc5a-Deficient Murine Lung Adenocarcinoma With Somatic Kras Mutations.Front Oncol. 2019 Apr 2;9:207. doi: 10.3389/fonc.2019.00207. eCollection 2019. Front Oncol. 2019. PMID: 31001473 Free PMC article.
-
Comparative functional genomics analysis of NNK tobacco-carcinogen induced lung adenocarcinoma development in Gprc5a-knockout mice.PLoS One. 2010 Jul 29;5(7):e11847. doi: 10.1371/journal.pone.0011847. PLoS One. 2010. PMID: 20686609 Free PMC article.
-
The mutational landscapes of genetic and chemical models of Kras-driven lung cancer.Nature. 2015 Jan 22;517(7535):489-92. doi: 10.1038/nature13898. Epub 2014 Nov 2. Nature. 2015. PMID: 25363767 Free PMC article.
-
Molecular pathogenesis of transplacentally induced mouse lung tumors.Exp Lung Res. 1998 Jul-Aug;24(4):557-77. doi: 10.3109/01902149809087386. Exp Lung Res. 1998. PMID: 9659583 Review.
-
Modeling K-Ras-driven lung adenocarcinoma in mice: preclinical validation of therapeutic targets.J Mol Med (Berl). 2016 Feb;94(2):121-35. doi: 10.1007/s00109-015-1360-5. Epub 2015 Nov 3. J Mol Med (Berl). 2016. PMID: 26526121 Review.
Cited by
-
Premalignant Progression in the Lung: Knowledge Gaps and Novel Opportunities for Interception of Non-Small Cell Lung Cancer. An Official American Thoracic Society Research Statement.Am J Respir Crit Care Med. 2024 Sep 1;210(5):548-571. doi: 10.1164/rccm.202406-1168ST. Am J Respir Crit Care Med. 2024. PMID: 39115548 Free PMC article. Review.
-
Crystalline silica particles cause rapid NLRP3-dependent mitochondrial depolarization and DNA damage in airway epithelial cells.Part Fibre Toxicol. 2020 Aug 10;17(1):39. doi: 10.1186/s12989-020-00370-2. Part Fibre Toxicol. 2020. PMID: 32778128 Free PMC article.
-
microRNA-367-3p regulation of GPRC5A is suppressed in ischemic stroke.J Cereb Blood Flow Metab. 2020 Jun;40(6):1300-1315. doi: 10.1177/0271678X19858637. Epub 2019 Jul 11. J Cereb Blood Flow Metab. 2020. PMID: 31296130 Free PMC article.
-
Genome-Wide and Phenotypic Evaluation of Stem Cell Progenitors Derived From Gprc5a-Deficient Murine Lung Adenocarcinoma With Somatic Kras Mutations.Front Oncol. 2019 Apr 2;9:207. doi: 10.3389/fonc.2019.00207. eCollection 2019. Front Oncol. 2019. PMID: 31001473 Free PMC article.
-
Alveolar type I cells can give rise to KRAS-induced lung adenocarcinoma.Cell Rep. 2023 Dec 26;42(12):113286. doi: 10.1016/j.celrep.2023.113286. Epub 2023 Nov 22. Cell Rep. 2023. PMID: 37995179 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. - PubMed
-
- Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2016;25:16–27. - PubMed
-
- Kadara H, Scheet P, Wistuba II, Spira AE. Early Events in the Molecular Pathogenesis of Lung Cancer. Cancer Prev Res (Phila) 2016;9:518–27. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

