Disentangling the effects of selection and loss bias on gene dynamics
- PMID: 28652353
- PMCID: PMC5514749
- DOI: 10.1073/pnas.1704925114
Disentangling the effects of selection and loss bias on gene dynamics
Abstract
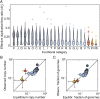
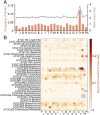
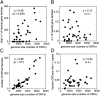
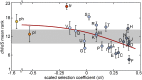
We combine mathematical modeling of genome evolution with comparative analysis of prokaryotic genomes to estimate the relative contributions of selection and intrinsic loss bias to the evolution of different functional classes of genes and mobile genetic elements (MGE). An exact solution for the dynamics of gene family size was obtained under a linear duplication-transfer-loss model with selection. With the exception of genes involved in information processing, particularly translation, which are maintained by strong selection, the average selection coefficient for most nonparasitic genes is low albeit positive, compatible with observed positive correlation between genome size and effective population size. Free-living microbes evolve under stronger selection for gene retention than parasites. Different classes of MGE show a broad range of fitness effects, from the nearly neutral transposons to prophages, which are actively eliminated by selection. Genes involved in antiparasite defense, on average, incur a fitness cost to the host that is at least as high as the cost of plasmids. This cost is probably due to the adverse effects of autoimmunity and curtailment of horizontal gene transfer caused by the defense systems and selfish behavior of some of these systems, such as toxin-antitoxin and restriction modification modules. Transposons follow a biphasic dynamics, with bursts of gene proliferation followed by decay in the copy number that is quantitatively captured by the model. The horizontal gene transfer to loss ratio, but not duplication to loss ratio, correlates with genome size, potentially explaining increased abundance of neutral and costly elements in larger genomes.
Keywords: antiparasite defense; gene loss; horizontal gene transfer; mobile genetic elements; selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Phylogenomics of Cas4 family nucleases.BMC Evol Biol. 2017 Nov 28;17(1):232. doi: 10.1186/s12862-017-1081-1. BMC Evol Biol. 2017. PMID: 29179671 Free PMC article.
-
Inevitability of Genetic Parasites.Genome Biol Evol. 2016 Sep 26;8(9):2856-2869. doi: 10.1093/gbe/evw193. Genome Biol Evol. 2016. PMID: 27503291 Free PMC article.
-
Causes of insertion sequences abundance in prokaryotic genomes.Mol Biol Evol. 2007 Apr;24(4):969-81. doi: 10.1093/molbev/msm014. Epub 2007 Jan 23. Mol Biol Evol. 2007. PMID: 17251179
-
Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review.
-
Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution.Nucleic Acids Res. 2001 Sep 15;29(18):3742-56. doi: 10.1093/nar/29.18.3742. Nucleic Acids Res. 2001. PMID: 11557807 Free PMC article. Review.
Cited by
-
The population genetics of prokaryotic pangenomes.Nat Ecol Evol. 2024 Feb;8(2):190-191. doi: 10.1038/s41559-023-02276-6. Nat Ecol Evol. 2024. PMID: 38177691 No abstract available.
-
Thermodynamics of evolution and the origin of life.Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2120042119. doi: 10.1073/pnas.2120042119. Proc Natl Acad Sci U S A. 2022. PMID: 35131858 Free PMC article.
-
On the networked architecture of genotype spaces and its critical effects on molecular evolution.Open Biol. 2018 Jul;8(7):180069. doi: 10.1098/rsob.180069. Open Biol. 2018. PMID: 29973397 Free PMC article. Review.
-
Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire.Nat Rev Genet. 2020 Feb;21(2):119-131. doi: 10.1038/s41576-019-0172-9. Epub 2019 Oct 14. Nat Rev Genet. 2020. PMID: 31611667 Review.
-
Selfish, promiscuous and sometimes useful: how mobile genetic elements drive horizontal gene transfer in microbial populations.Philos Trans R Soc Lond B Biol Sci. 2022 Oct 10;377(1861):20210234. doi: 10.1098/rstb.2021.0234. Epub 2022 Aug 22. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35989606 Free PMC article. Review.
References
-
- Koonin EV. The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press; Upper Saddle River, NJ: 2011.
-
- Lynch M. The Origins of Genome Architecture. Sinauer Associates; Sunderland, MA: 2007.
-
- Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

