Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation
- PMID: 28645917
- PMCID: PMC5538766
- DOI: 10.15252/embj.201695757
Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation
Abstract
Replacement of canonical histones with specialized histone variants promotes altering of chromatin structure and function. The essential histone variant H2A.Z affects various DNA-based processes via poorly understood mechanisms. Here, we determine the comprehensive interactome of H2A.Z and identify PWWP2A as a novel H2A.Z-nucleosome binder. PWWP2A is a functionally uncharacterized, vertebrate-specific protein that binds very tightly to chromatin through a concerted multivalent binding mode. Two internal protein regions mediate H2A.Z-specificity and nucleosome interaction, whereas the PWWP domain exhibits direct DNA binding. Genome-wide mapping reveals that PWWP2A binds selectively to H2A.Z-containing nucleosomes with strong preference for promoters of highly transcribed genes. In human cells, its depletion affects gene expression and impairs proliferation via a mitotic delay. While PWWP2A does not influence H2A.Z occupancy, the C-terminal tail of H2A.Z is one important mediator to recruit PWWP2A to chromatin. Knockdown of PWWP2A in Xenopus results in severe cranial facial defects, arising from neural crest cell differentiation and migration problems. Thus, PWWP2A is a novel H2A.Z-specific multivalent chromatin binder providing a surprising link between H2A.Z, chromosome segregation, and organ development.
Keywords: H2A.Z; PWWP2A; chromatin; mitosis; neural crest.
© 2017 The Authors.
Figures
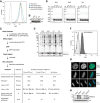
Flow cytometry analysis of HeLaK cells either non‐transfected (wt, gray histogram) or stably transfected with GFP‐H2A (green histogram), GFP‐H2A.Z.1 (blue histogram), and GFP‐H2A.Z.2 (orange histogram). Notice that expression levels of tagged histones are similar.
Immunoblots of nuclear extracts derived from HeLaK cells stably expressing GFP‐tagged histones (see also A). Anti‐H3 antibody was used as loading control.
Schematic workflow of LFQ‐MS experiment (see also Fig 1A and Materials and Methods). Table depicts the number of proteins identified by indicated method in each step of the procedure.
Coomassie staining of proteins found in S1, S2, and pellet (P) fractions after MNase digest of chromatin derived from SKmel‐147 cells stably expressing GFP, GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.2), or GFP‐H2A.Z.2 (Z.2). Notice that all fractions contain histones, but remaining protein content is different.
Bioanalyzer separated DNA sizes of S1‐containing nucleosomes after MNase digest of two independent SKmel‐147 replicates (SK1 and SK2) or HeLaK (HK) cells stably expressing GFP, GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.2), or GFP‐H2A.Z.2 (Z.2) used in LFQ‐MS experiments shown in Fig 1A. Notice that S1 fractions that were used for LFQ‐MS IP experiments contain > 90% pure mononucleosomes as seen by DNA sizes of around 150 bp.
Flow cytometry analysis of HeLaK cells either non‐transfected (control, light gray histogram) or stably transfected with GFP‐PWWP2A (dark gray histogram). Notice that a pure population of HeLaK cells expressing GFP‐PWWP2A at low level was obtained.
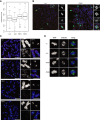
Confocal microscopy images of HeLaK cells stably expressing GFP‐PWWP2A in interphase, metaphase, or anaphase. DNA was visualized by DAPI staining (blue), and GFP‐PWWP2A is seen in green. Scale bars = 10 μm. Notice that some GFP‐PWWP2A remains stably associated with condensed mitotic chromatin.
Immunoblotting of endogenous PWWP2A upon mono‐IPs using antibodies against endogenous H2A or H2A.Z. Arrow indicates endogenous PWWP2A.

- A
Heatmap of significant outliers from pull‐downs analyzed by label‐free MS‐based proteomics with mononucleosomes derived from Skmel‐147 (two independent experiments: SK1 and SK2; see also Vardabasso et al, 2015 for first replicate) or HeLaK (HK) cells stably expressing GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.1), or GFP‐H2A.Z.2 (Z.2), normalized to H2A. Scale bar: log2‐fold. See also Fig EV1A–E for details on experimental procedure and verification of results, and Datasets EV1, EV2 and EV3 for detailed lists of H2A.Z‐binders.
- B
Immunoblotting of SRCAP complex‐specific member ZNHIT1 upon SK‐mel147 GFP, GFP‐H2A, GFP‐H2A.Z.1, or GFP‐H2A.Z.2 mono‐IPs. GFP served as control.
- C, D
Immunoblotting of different histone PTMs (C) or PWWP2A (D) upon GFP, GFP‐H2A, GFP‐H2A.Z.1, or GFP‐H2A.Z.2 mono‐IPs. Notice the different sizes of endogenous PWWP2A protein (see also Figs EV1H and EV4C), possibly due to different modifications.
- E
Immunoblots of GFP‐H2A.Z.1 or GFP‐PWWP2A mono‐IPs detecting endogenous H3, H2A, or H2A.Z. Notice enrichment of H2A.Z in comparison with H2A in GFP‐PWWP2A pull‐down. See also Fig EV1F–H for generation and characterization of GFP‐PWWP2A cell lines and endogenous pull‐down.

Pull‐downs of GST or GST‐PWWP2A with mononucleosomes (input) derived from HeLaK cells. Precipitated recombinant GST proteins and histones are detected with Coomassie blue staining and H2A and H2A.Z with specific antibodies in immunoblots. * indicates right sizes of purified and precipitated GST and GST‐PWWP2A.
Immunoblots of GST‐PWWP2A IPs with mononucleosomes derived from HeLaK cells stably expressing GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.1), or GFP‐H2A.Z.2 (Z.2).
Representative competitive EMSA using recombinant H2A (bottom)‐ or H2A.Z (top)‐containing nucleosomes incubated with indicated increasing concentrations of GST‐PWWP2A. GST alone served as negative control. * indicates nucleosome; ** indicates nucleosome‐GST‐PWWP2A complex.
Schematic representation of recombinant GST‐tagged PWWP2A and PWWP domain deletion (ΔPWWP) and PWWP domain only (PWWP)‐containing constructs (top) used in cell‐derived mono‐IPs followed by Coomassie staining and immunoblotting (bottom). * indicates respective GST proteins. Notice that both the PWWP domain alone as well as the PWWP‐deletion protein are able to interact with nucleosomes, indicating at least two independent nucleosome binding sites within PWWP2A.
Schematic representation of recombinant GST‐PWWP2A deletions (top) used in cell‐derived mono‐IPs followed by Coomassie and immunoblotting (bottom). See also Appendix Fig S1B for protein purification.
Schematic representation of recombinant GST‐PWWP2A internal deletions (top) used in cell‐derived mono‐IPs followed by Coomassie and immunoblotting (bottom). See also Appendix Fig S1C and D for protein purification and further IPs.
IPs as described in (F) with mononucleosomes derived from HeLaK cells stably expressing GFP‐H2A and GFP‐H2A.Z isoforms detected with anti‐GFP antibody.
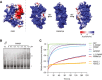
The electrostatic surface potential (ESP) of the published structure of the PSIP DNA‐binding PWWP domain (Eidahl et al, 2013) was calculated (left), and the PWWP2A‐PWWP domain (right) was modeled based on published PWWP2B 3D structure (Qin & Min, 2014). ESP color values are in units of kcal/(mol*e) at 298 K. Three representative views of the ESP of the PWWP2A PWWP domain (right).
Representative EMSA using Cy‐5 labeled 75‐bp dsDNA and indicated increasing concentrations of GST‐PWWP domain. *: free DNA; **: shifted PWWP‐DNA complex. See also Appendix Fig S2A for H2A.Z‐independence of PWWP domain in nucleosome interaction.
FRAP quantification curves of average GFP signal relative to fluorescence signal prior to bleaching from transiently expressed GFP (negative control), GFP‐tagged histones, and GFP‐tagged PWWP2A mutants (n = 5–14). See also Appendix Fig S2B for FRAP experiments comparing recovery signals of different chromatin binding proteins to GFP‐PWWP2A, as well as Appendix Fig S2C–E for FRAP analyses of stable and transient GFP and GFP‐tagged proteins expressing HeLaK cells.
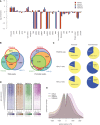
Log2‐enrichment plot representing genomic regions after GFP‐PWWP2A (blue), GFP‐H2A.Z.1 (red), GFP‐H2A.Z.2 (orange), and H3K4me3 (purple) nChIP‐seq. Shown are two biological replicates for each nChIP. See also Fig EV2A for representative genome browser captures.
Venn diagrams displaying total (left) or promoter‐occupying (right) peaks of HeLaK GFP‐PWWP2A, GFP‐H2A.Z.1, and GFP‐H2A.Z.2. Promoters are defined as −3 kb < TSS < +1 kb.
Distribution of overlapping peaks between GFP‐PWWP2A (top), GFP‐H2A.Z.1 (middle), and GFP‐H2A.Z.2 (bottom) nChIP‐seq data of promoter (left) and non‐promoter (right) regions according to (B).
Heatmap of nChIP‐seq peaks at transcriptional start sites (TSS) sorted for expression level (top: high expressed, bottom: low expressed genes).
Correlation of GFP‐PWWP2A (green), ‐H2A.Z.1 (blue), ‐H2A.Z.2 (brown), and H3K4me3 (purple) mean coverage signals at TSS of expressed genes. See Fig EV2B for GFP‐PWWP2A localization at euchromatic regions.

Genome browser snap shot of a representative region in chromosome 20 displaying two independent replicates of HeLaK GFP‐H2A.Z.1, GFP‐H2A.Z.2, GFP‐PWWP2A, and H3K4me3 nChIPs signals at intergenic, gene body, and TSS regions. Annotated genes are displayed below. See also Fig 4 for nChIP‐seq analyses.
Representative confocal IF pictures of HeLaK cells stably expressing GFP‐PWWP2A (green), co‐stained with DAPI (blue) and either H3K4me3 antibody (top, red) or H3K9me3 (bottom, red). Scale bars = 10 μm.
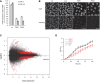
PWWP2A expression analysis by qPCR 2 days after knockdown with two independent siRNAs (PW#1, PW#2) using two different primer pairs (PW_2: recognizes main splice product; PW_all: recognizes all predicted splice forms). Luciferase siRNA (Luci) and non‐transfected wild‐type (wt) cells were used as controls. All data were normalized to HPRT1/HMBSS expression and depicted as % of luci transfectants. Error bars indicate SEM of four biological replicates.
IF microscopy analysis of cells treated as described in (A). Cells were stained with DAPI to visualize DNA (top) and anti‐PWWP2A antibody (bottom). Scale bars = 10 μm. Notice slight increase in nuclear size upon PWWP2A depletion (quantification in Fig EV3A).
Scatter plot summarizing genes up‐ (top red) or down‐regulated (bottom red) upon PWWP2A depletion (blue dots indicate different PWWP2A RNA isoforms) as determined by RNA‐seq. See also Appendix Fig S3 for GO term analysis of deregulated transcripts.
Growth curve of HeLaK cells after RNAi (red: PWWP2A siRNAs, black: wt and luciferase control siRNAs). Error bars indicate SEM of three independent biological replicates.
Standard Tukey boxplots of nuclear sizes of control (ctrlii, luci) and PWWP2A‐depleted (PW#1, PW#2) cells (n = 3). Horizontal line = median, box = interquartile range.
Confocal IF microscopy analysis of HeLaK RNAi cells to visualize mitotic phases. Two days after RNAi cells were stained with DAPI (blue), H3S10ph (green), and tubulin (red) antibodies. Scale bars = 10 μm. See also Fig 6E for quantitation of mitotic stages.
Representative confocal IF pictures of chromosomes of control (ctrlii, Luci) and PWWP2A‐depleted (PW#1, PW#2) HeLaK cells co‐stained with DAPI (blue), CENP‐E (green), and PWWP2A (red). Scale bars = 10 μm (enlarged chromosomes in boxes; scale bars = 5 μm).
Representative confocal IF pictures of mitotic HeLaK cells two days after control (wt, Luci) and PWWP2A (PW#1, PW#2) siRNA transfections. HeLaK cells were co‐stained with DAPI (blue) and tubulin (green) antibodies. Scale bars = 10 μm.

Cell cycle analysis of PI‐stained HeLaK RNAi cells using flow cytometer.
Quantification of experiments described in (A). Percentages of cells in G1, S, or G2/M phases 3 days after RNAi. Error bars indicate SEM of five independent biological replicates.
Flow cytometry dot‐plots of HeLaK cells co‐stained with PI and H3S10ph antibody 3 days after RNAi to distinguish between G2 and M phase cells.
Quantification of experiments described in (C). Percentages of cells in G2 or M phases 3 days after RNAi. Error bars indicate SEM of three independent biological replicates.
Quantification of mitotic phases 2 days after RNAi by visually distinguishing morphological characteristics of H3S10ph‐positive cells. Error bars indicate SEM of four independent biological replicates. See also Fig EV3B–D for stainings and chromosome spreads of control and RNAi cells.
Selected panels from live cell time‐lapse experiments (see also Movies EV1, EV2, EV3 and EV4) of wt, luciferase control, and PWWP2A‐depleted HeLaK cells expressing GFP‐H2A for visualization of chromatin for 4 h starting from nuclear breakdown at prophase. Scale bars = 10 μm.
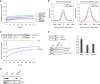
FRAP quantification curves of average GFP signal relative to fluorescence signal prior to bleaching from HeLaK stably expressing GFP‐H2A or GFP‐H2A.Z.1 2 days after control (wt, Luci) or PWWP2A (PW#1, PW#2) knockdown (n = 9–19) (see Appendix Fig S4A for FRAP experiments).
Venn diagram displaying sole GFP‐H2A.Z.1 (left, 22,343 peaks) or overlapping with PWWP2A (left, 18,227 peaks) nChIP‐seq signals in control (ctrlii, Luci; dark blue) or PWWP2A‐depleted (PW#1, PW#2; red) background (see also Fig EV4A and B).
Schematic representation of PWWP2A and IC region deletion (ΔIC) (top). FRAP quantification curves (bottom) as shown and described in Fig 3C with the addition of GFP‐ΔIC (n = 16) (see also Appendix Fig S4C for FRAP IF pictures).
Immunoblotting of endogenous H3 and H2A.Z (verification of tetracycline (tet)‐induced knockout efficiency) upon IP of mononucleosomes derived from WT or H2A.Z DKO DT40 cells with recombinant GST (negative control) or GST‐PWWP2A. Notice that less H3 is pulled down with PWWP2A when H2A.Z is depleted.
Schematic representation of Flag‐tagged chicken H2A, H2A.Z, and H2A.Z C‐terminal deletion mutant (H2A.ZΔC) (top, left). Note indication of acidic patch in H2A (gray box) and extended acidic patch in H2A.Z (black box). After transient transfection of HK cells with Flag‐constructs, derived mononucleosomes were incubated with recombinant GST or GST‐PWWP2A and binding efficiency and variant‐specificity was tested in immunoblots with anti‐Flag antibody (left). Right: Quantification of signal intensities of immunoblots using Image Studio Lite Ver 5.2 (LI‐COR). Error bars indicate SEM of four independent replicates. See Fig EV4E for partial growth rescue of Flag‐H2A.ZΔC in DKO cells.
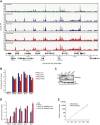
Genome browser snap shot of a representative region in chromosome 7 displaying GFP‐PWWP2A (green, taken from Fig 4) and GFP‐H2A.Z nChIP‐seq signals 2 days after control (ctrlii, Luci, blue) or PWWP2A depletion (PW#1, PW#2, red). Annotated gene structures are shown below. Notice that H2A.Z peaks do not significantly change upon PWWP2A knockdown and in comparison with PWWP2A‐containing or PWWP2A‐missing sites. See also Fig 7B for Venn diagrams.
nChIP‐qPCR of GFP‐H2A.Z to verify results obtained in (A) and Fig 7B. RPL11 represents a gene body site missing H2A.Z nucleosomes (negative control), while all other regions show H2A.Z‐containing promoter sites (− = −1 nucleosome, + = +1 nucleosome). Error bars indicate SEM of two to three technical replicates. Notice that PWWP2A depletion does not lead to a significant change in H2A.Z occupancy.
Immunoblot of cell extracts 2 days after control (wt, Luci) or PWWP2A (PW#1, PW#2) knockdown with antibodies against PWWP2A, H3, and H2A.Z. Notice that PWWP2A reduction does not influence global H2A.Z levels.
nChIP‐qPCR of GFP (negative control, light gray), GFP‐H2A.Z without exogenous Cherry‐PWWP2A (Ch‐PWWP2A) expression (dark gray) or upon sorting of low (light red) or high (dark red) Ch‐PWWP2A expression levels (see Appendix Fig S4B for FACS levels) using primer pairs shown in Fig EV4B. Error bars indicate SEM of two to three technical replicates. Notice that PWWP2A overexpression does not significantly affect H2A.Z occupancy.
Growth curve of DKO cells rescued with either wt Flag‐H2A.Z (red, see also Kusakabe et al, 2016) or Flag‐H2A.ZΔC (green). Notice that although Flag‐H2A.ZΔC transfected cells grow slower than cells expressing wt Flag‐H2A.Z, they are able to partially rescue the severe mitotic defect observed in DKO cells implying chromatin deposition took place (see also Fig 7E).

Expression pattern of endogenous Xenopus tropicalis pwwp2a mRNA during early development. Pictures show whole‐mount RNA in situ hybridization patterns (purple color) for the following developmental stages: egg, blastula and early (e‐)/late (l‐) gastrula stages in lateral views of sagittal sections; e‐neurula and e‐tailbud stages show anterior views (left picture) or dorsal views (right picture, anterior to the left). Mid (m‐) and l‐tailbud stages are shown in lateral views (anterior left). Abbreviations: ba, branchial arches; cnc, cranial neural crest; eye, retinal Anlage; nf, neural folds; ov, otic vesicle.
Top: Targeting region of the pwMO oligonucleotide on the X. laevis pwwp2a mRNA. The translational start site is highlighted in red. To determine the translational blocking efficiency of pwMO in vivo, a ˜275‐bp fragment of pwwp2a 5′ cDNA sequence, including the AUG, was cloned in frame upstream of the luciferase coding region. Bottom: The above depicted luciferase construct was injected in CoMO‐ or pwMO‐loaded embryos and chemiluminescence measured at gastrula stage. Error bars indicate SEM of three independent biological replicates. UI, uninjected embryos.
Morphology of X. tropicalis embryos injected with CoMO and pwMO together with lineage‐tracer Alexa‐488; numbers indicate penetrance of major morphological phenotype over total embryos inspected (n = 3 experiments).

Left side: One‐cell of two‐cell stage Xenopus laevis embryos was injected with either control (CoMO) or pwwp2a‐specific (pwMO) morpholino. For rescue experiments, pwMO morphant embryos were coinjected with mRNAs encoding either GFP‐tagged full‐length or variant human PWWP2A proteins, as indicated. Injected body sites were identified by Alexa 594 red fluorescence, while GFP fluorescence indicates synthesis of coinjected human PWWP2A protein variants. Panels display side views of representative embryos from the indicated conditions; numbers indicate penetrance of the major morphological phenotype over total embryos inspected. Right side: Quantification of the percentage of the observed phenotype, that is, malformation of head structures and eyes. Error bars indicate SEM of at least three independent biological replicates (see also Table EV1). *P ≤ 0.05 (Student's t‐test, two‐sided, unpaired).
Whole‐mount RNA in situ hybridization assays with probes against rx1 (anterior view) and twist (dorsoanterior view) in either CoMO‐ or pwMO‐injected embryos at neurula stage. The rx1 gene is induced normally; the cranial neural crest marker twist is diminished on the pwMO‐injected side (91% affected), which is partially restored by coinjection of full‐length PWWP2A mRNA (60% affected). Numbers indicate penetrance of the depicted molecular phenotype over total embryos inspected. Right: Quantification of the percentage of misexpression of twist mRNA in controls compared to pwMO morphant and rescue condition. Error bars indicate SEM of three independent biological replicates. *P ≤ 0.05 (Student's t‐test, two‐sided, unpaired).
Representative images of dissected facial cartilage from CoMO‐ or pwMO‐injected embryos visualized by Alcian blue staining. (+) indicates injected body side.
Comment in
-
PWWP2A: A novel mitosis link?Cell Cycle. 2017 Oct 18;16(20):1849-1850. doi: 10.1080/15384101.2017.1372523. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933988 Free PMC article. No abstract available.
Similar articles
-
The H2A.Z and NuRD associated protein HMG20A controls early head and heart developmental transcription programs.Nat Commun. 2023 Jan 28;14(1):472. doi: 10.1038/s41467-023-36114-x. Nat Commun. 2023. PMID: 36709316 Free PMC article.
-
PWWP2A binds distinct chromatin moieties and interacts with an MTA1-specific core NuRD complex.Nat Commun. 2018 Oct 16;9(1):4300. doi: 10.1038/s41467-018-06665-5. Nat Commun. 2018. PMID: 30327463 Free PMC article.
-
The histone variant H2A.Z is an important regulator of enhancer activity.Nucleic Acids Res. 2015 Nov 16;43(20):9742-56. doi: 10.1093/nar/gkv825. Epub 2015 Aug 28. Nucleic Acids Res. 2015. PMID: 26319018 Free PMC article.
-
Patterning chromatin: form and function for H2A.Z variant nucleosomes.Curr Opin Genet Dev. 2006 Apr;16(2):119-24. doi: 10.1016/j.gde.2006.02.005. Epub 2006 Feb 28. Curr Opin Genet Dev. 2006. PMID: 16503125 Review.
-
Regulation of gene expression and cellular proliferation by histone H2A.Z.Biochem Cell Biol. 2009 Feb;87(1):179-88. doi: 10.1139/O08-138. Biochem Cell Biol. 2009. PMID: 19234533 Review.
Cited by
-
The H2A.Z and NuRD associated protein HMG20A controls early head and heart developmental transcription programs.Nat Commun. 2023 Jan 28;14(1):472. doi: 10.1038/s41467-023-36114-x. Nat Commun. 2023. PMID: 36709316 Free PMC article.
-
JASPer controls interphase histone H3S10 phosphorylation by chromosomal kinase JIL-1 in Drosophila.Nat Commun. 2019 Nov 25;10(1):5343. doi: 10.1038/s41467-019-13174-6. Nat Commun. 2019. PMID: 31767855 Free PMC article.
-
Spatial organization of transcribed eukaryotic genes.Nat Cell Biol. 2022 Mar;24(3):327-339. doi: 10.1038/s41556-022-00847-6. Epub 2022 Feb 17. Nat Cell Biol. 2022. PMID: 35177821 Free PMC article.
-
A de novo SFMBT1 pathogenic variant identified in a boy with Poland syndrome.Cold Spring Harb Mol Case Stud. 2022 Apr 28;8(3):a006168. doi: 10.1101/mcs.a006168. Print 2022 Apr. Cold Spring Harb Mol Case Stud. 2022. PMID: 35483874 Free PMC article.
-
The Role of the Histone Variant H2A.Z in Metazoan Development.J Dev Biol. 2022 Jul 1;10(3):28. doi: 10.3390/jdb10030028. J Dev Biol. 2022. PMID: 35893123 Free PMC article. Review.
References
-
- Auckland P, McAinsh AD (2015) Building an integrated model of chromosome congression. J Cell Sci 128: 3363–3374 - PubMed
-
- Bellmeyer A, Krase J, Lindgren J, LaBonne C (2003) The protooncogene c‐myc is an essential regulator of neural crest formation in xenopus. Dev Cell 4: 827–839 - PubMed
-
- Bonisch C, Schneider K, Punzeler S, Wiedemann SM, Bielmeier C, Bocola M, Eberl HC, Kuegel W, Neumann J, Kremmer E, Leonhardt H, Mann M, Michaelis J, Schermelleh L, Hake SB (2012) H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res 40: 5951–5964 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

