Interplay of PA-X and NS1 Proteins in Replication and Pathogenesis of a Temperature-Sensitive 2009 Pandemic H1N1 Influenza A Virus
- PMID: 28637750
- PMCID: PMC5553184
- DOI: 10.1128/JVI.00720-17
Interplay of PA-X and NS1 Proteins in Replication and Pathogenesis of a Temperature-Sensitive 2009 Pandemic H1N1 Influenza A Virus
Abstract
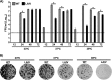
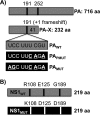
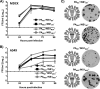
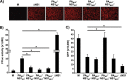
Influenza A viruses (IAVs) cause seasonal epidemics and occasional pandemics, representing a serious public health concern. It has been described that one mechanism used by some IAV strains to escape the host innate immune responses and modulate virus pathogenicity involves the ability of the PA-X and NS1 proteins to inhibit the host protein synthesis in infected cells. It was reported that for the 2009 pandemic H1N1 IAV (pH1N1) only the PA-X protein had this inhibiting capability, while the NS1 protein did not. In this work, we have evaluated, for the first time, the combined effect of PA-X- and NS1-mediated inhibition of general gene expression on virus pathogenesis, using a temperature-sensitive, live-attenuated 2009 pandemic H1N1 IAV (pH1N1 LAIV). We found that viruses containing PA-X and NS1 proteins that simultaneously have (PAWT+/NS1MUT+) or do not have (PAMUT-/NS1WT-) the ability to block host gene expression showed reduced pathogenicity in vivo However, a virus where the ability to inhibit host protein expression was switched between PA-X and NS1 (PAMUT-/NS1MUT+) presented pathogenicity similar to that of a virus containing both wild-type proteins (PAWT+/NS1WT-). Our findings suggest that inhibition of host protein expression is subject to a strict balance, which can determine the successful progression of IAV infection. Importantly, knowledge obtained from our studies could be used for the development of new and more effective vaccine approaches against IAV.IMPORTANCE Influenza A viruses (IAVs) are one of the most common causes of respiratory infections in humans, resulting in thousands of deaths annually. Furthermore, IAVs can cause unpredictable pandemics of great consequence when viruses not previously circulating in humans are introduced into humans. The defense machinery provided by the host innate immune system limits IAV replication; however, to counteract host antiviral activities, IAVs have developed different inhibition mechanisms, including prevention of host gene expression mediated by the viral PA-X and NS1 proteins. Here, we provide evidence demonstrating that optimal control of host protein synthesis by IAV PA-X and/or NS1 proteins is required for efficient IAV replication in the host. Moreover, we demonstrate the feasibility of genetically controlling the ability of IAV PA-X and NS1 proteins to inhibit host immune responses, providing an approach to develop more effective vaccines to combat disease caused by this important respiratory pathogen.
Keywords: host response; influenza; influenza vaccines; virus-host interactions.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
Functional Evolution of the 2009 Pandemic H1N1 Influenza Virus NS1 and PA in Humans.J Virol. 2018 Sep 12;92(19):e01206-18. doi: 10.1128/JVI.01206-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021892 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Functional Evolution of Influenza Virus NS1 Protein in Currently Circulating Human 2009 Pandemic H1N1 Viruses.J Virol. 2017 Aug 10;91(17):e00721-17. doi: 10.1128/JVI.00721-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637754 Free PMC article.
-
Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza A Viruses Carrying Human or Avian Nonstructural Protein 1.J Virol. 2019 Dec 12;94(1):e00999-19. doi: 10.1128/JVI.00999-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597767 Free PMC article.
-
Influenza Virus: A Master Tactician in Innate Immune Evasion and Novel Therapeutic Interventions.Front Immunol. 2018 Apr 12;9:743. doi: 10.3389/fimmu.2018.00743. eCollection 2018. Front Immunol. 2018. PMID: 29755452 Free PMC article. Review.
Cited by
-
Antiviral responses versus virus-induced cellular shutoff: a game of thrones between influenza A virus NS1 and SARS-CoV-2 Nsp1.Front Cell Infect Microbiol. 2024 Feb 5;14:1357866. doi: 10.3389/fcimb.2024.1357866. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38375361 Free PMC article. Review.
-
Zoonosis and zooanthroponosis of emerging respiratory viruses.Front Cell Infect Microbiol. 2024 Jan 5;13:1232772. doi: 10.3389/fcimb.2023.1232772. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38249300 Free PMC article. Review.
-
NS1 and PA-X of H1N1/09 influenza virus act in a concerted manner to manipulate the innate immune response of porcine respiratory epithelial cells.Front Cell Infect Microbiol. 2023 Jul 26;13:1222805. doi: 10.3389/fcimb.2023.1222805. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37565063 Free PMC article.
-
Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs.Pathogens. 2022 Dec 12;11(12):1521. doi: 10.3390/pathogens11121521. Pathogens. 2022. PMID: 36558855 Free PMC article. Review.
-
PA-X protein of H1N1 subtype influenza virus disables the nasal mucosal dendritic cells for strengthening virulence.Virulence. 2022 Dec;13(1):1928-1942. doi: 10.1080/21505594.2022.2139474. Virulence. 2022. PMID: 36271710 Free PMC article.
References
-
- Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA (ed), Fields virology, 5th ed Lippincott Williams and Wilkins, Philadelphia, PA.
-
- Chan W, Zhou H, Kemble G, Jin H. 2008. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology 380:304–311. doi:10.1016/j.virol.2008.07.027. - DOI - PubMed
-
- Cox NJ, Kitame F, Kendal AP, Maassab HF, Naeve C. 1988. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology 167:554–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

