Stacks off tracks: a role for the golgin AtCASP in plant endoplasmic reticulum-Golgi apparatus tethering
- PMID: 28605454
- PMCID: PMC5853478
- DOI: 10.1093/jxb/erx167
Stacks off tracks: a role for the golgin AtCASP in plant endoplasmic reticulum-Golgi apparatus tethering
Abstract
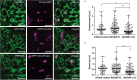
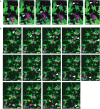
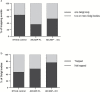
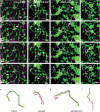
The plant Golgi apparatus modifies and sorts incoming proteins from the endoplasmic reticulum (ER) and synthesizes cell wall matrix material. Plant cells possess numerous motile Golgi bodies, which are connected to the ER by yet to be identified tethering factors. Previous studies indicated a role for cis-Golgi plant golgins, which are long coiled-coil domain proteins anchored to Golgi membranes, in Golgi biogenesis. Here we show a tethering role for the golgin AtCASP at the ER-Golgi interface. Using live-cell imaging, Golgi body dynamics were compared in Arabidopsis thaliana leaf epidermal cells expressing fluorescently tagged AtCASP, a truncated AtCASP-ΔCC lacking the coiled-coil domains, and the Golgi marker STtmd. Golgi body speed and displacement were significantly reduced in AtCASP-ΔCC lines. Using a dual-colour optical trapping system and a TIRF-tweezer system, individual Golgi bodies were captured in planta. Golgi bodies in AtCASP-ΔCC lines were easier to trap and the ER-Golgi connection was more easily disrupted. Occasionally, the ER tubule followed a trapped Golgi body with a gap, indicating the presence of other tethering factors. Our work confirms that the intimate ER-Golgi association can be disrupted or weakened by expression of truncated AtCASP-ΔCC and suggests that this connection is most likely maintained by a golgin-mediated tethering complex.
Keywords: Arabidopsis; Golgi apparatus; endomembrane system; endoplasmic reticulum; golgin; optical tweezers; secretory pathway; tethering factor.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

Comment in
-
Endoplasmic reticulum and Golgi apparatus: old friends, novel intimate relationships.J Exp Bot. 2017 Jun 15;68(13):3283-3285. doi: 10.1093/jxb/erx216. J Exp Bot. 2017. PMID: 28859381 Free PMC article.
Similar articles
-
Living on the edge: the role of Atgolgin-84A at the plant ER-Golgi interface.J Microsc. 2020 Nov;280(2):158-173. doi: 10.1111/jmi.12946. Epub 2020 Jul 31. J Microsc. 2020. PMID: 32700322
-
Localization and domain characterization of Arabidopsis golgin candidates.J Exp Bot. 2007;58(15-16):4373-86. doi: 10.1093/jxb/erm304. J Exp Bot. 2007. PMID: 18182439
-
Identification and characterization of AtCASP, a plant transmembrane Golgi matrix protein.Plant Mol Biol. 2005 May;58(1):109-22. doi: 10.1007/s11103-005-4618-4. Plant Mol Biol. 2005. PMID: 16028120
-
Finding the Golgi: Golgin Coiled-Coil Proteins Show the Way.Trends Cell Biol. 2016 Jun;26(6):399-408. doi: 10.1016/j.tcb.2016.02.005. Epub 2016 Mar 11. Trends Cell Biol. 2016. PMID: 26972448 Review.
-
Golgins in the structure and dynamics of the Golgi apparatus.Curr Opin Cell Biol. 2003 Aug;15(4):405-13. doi: 10.1016/s0955-0674(03)00054-1. Curr Opin Cell Biol. 2003. PMID: 12892780 Review.
Cited by
-
Advances in Plant ER Architecture and Dynamics.Plant Physiol. 2018 Jan;176(1):178-186. doi: 10.1104/pp.17.01261. Epub 2017 Oct 6. Plant Physiol. 2018. PMID: 28986423 Free PMC article. Review.
-
Differentiation of Trafficking Pathways at Golgi Entry Core Compartments and Post-Golgi Subdomains.Front Plant Sci. 2020 Dec 8;11:609516. doi: 10.3389/fpls.2020.609516. eCollection 2020. Front Plant Sci. 2020. PMID: 33363561 Free PMC article. Review.
-
On the nature of the plant ER exit sites.Front Plant Sci. 2022 Oct 7;13:1010569. doi: 10.3389/fpls.2022.1010569. eCollection 2022. Front Plant Sci. 2022. PMID: 36275575 Free PMC article.
-
Genome-wide significant risk loci for mood disorders in the Old Order Amish founder population.Mol Psychiatry. 2023 Dec;28(12):5262-5271. doi: 10.1038/s41380-023-02014-1. Epub 2023 Mar 7. Mol Psychiatry. 2023. PMID: 36882501 Free PMC article.
-
An Update on the Key Factors Required for Plant Golgi Structure Maintenance.Front Plant Sci. 2022 Jun 28;13:933283. doi: 10.3389/fpls.2022.933283. eCollection 2022. Front Plant Sci. 2022. PMID: 35837464 Free PMC article. Review.
References
-
- Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. 2004. Molecular basis for Golgi maintenance and biogenesis. Current Opinion in Cell Biology 16, 364–372. - PubMed
-
- Barr FA, Short B. 2003. Golgins in the structure and dynamics of the Golgi apparatus. Current Opinion in Cell Biology 15, 405–413. - PubMed
-
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. 1998. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. The Plant Journal 15, 441–447. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

