Proteases as antimalarial targets: strategies for genetic, chemical, and therapeutic validation
- PMID: 28599096
- PMCID: PMC5575534
- DOI: 10.1111/febs.14130
Proteases as antimalarial targets: strategies for genetic, chemical, and therapeutic validation
Abstract
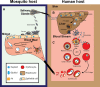
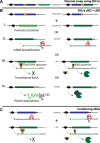
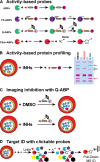
Malaria is a devastating parasitic disease affecting half of the world's population. The rapid emergence of resistance against new antimalarial drugs, including artemisinin-based therapies, has made the development of drugs with novel mechanisms of action extremely urgent. Proteases are enzymes proven to be well suited for target-based drug development due to our knowledge of their enzymatic mechanisms and active site structures. More importantly, Plasmodium proteases have been shown to be involved in a variety of pathways that are essential for parasite survival. However, pharmacological rather than target-based approaches have dominated the field of antimalarial drug development, in part due to the challenge of robustly validating Plasmodium targets at the genetic level. Fortunately, over the last few years there has been significant progress in the development of efficient genetic methods to modify the parasite, including several conditional approaches. This progress is finally allowing us not only to validate essential genes genetically, but also to study their molecular functions. In this review, I present our current understanding of the biological role proteases play in the malaria parasite life cycle. I also discuss how the recent advances in Plasmodium genetics, the improvement of protease-oriented chemical biology approaches, and the development of malaria-focused pharmacological assays, can be combined to achieve a robust biological, chemical and therapeutic validation of Plasmodium proteases as viable drug targets.
Keywords: malaria; protease; target validation.
© 2017 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures

Similar articles
-
Plasmodium falciparum proteases as new drug targets with special focus on metalloproteases.Mol Biochem Parasitol. 2024 Jun;258:111617. doi: 10.1016/j.molbiopara.2024.111617. Epub 2024 Mar 29. Mol Biochem Parasitol. 2024. PMID: 38554736 Review.
-
Targeting malaria protein kinases.Adv Protein Chem Struct Biol. 2021;124:225-274. doi: 10.1016/bs.apcsb.2020.10.004. Epub 2020 Nov 1. Adv Protein Chem Struct Biol. 2021. PMID: 33632466 Review.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
Plasmodium proteases and their role in development of Malaria vaccines.Adv Parasitol. 2024;126:253-273. doi: 10.1016/bs.apar.2024.08.001. Epub 2024 Sep 25. Adv Parasitol. 2024. PMID: 39448193
-
Protease-associated cellular networks in malaria parasite Plasmodium falciparum.BMC Genomics. 2011 Dec 23;12 Suppl 5(Suppl 5):S9. doi: 10.1186/1471-2164-12-S5-S9. Epub 2011 Dec 23. BMC Genomics. 2011. PMID: 22369208 Free PMC article.
Cited by
-
Egress and invasion machinery of malaria: an in-depth look into the structural and functional features of the flap dynamics of plasmepsin IX and X.RSC Adv. 2018 Jun 13;8(39):21829-21840. doi: 10.1039/c8ra04360d. eCollection 2018 Jun 13. RSC Adv. 2018. PMID: 35541758 Free PMC article.
-
Comparing sequence and structure of falcipains and human homologs at prodomain and catalytic active site for malarial peptide based inhibitor design.Malar J. 2019 May 3;18(1):159. doi: 10.1186/s12936-019-2790-2. Malar J. 2019. PMID: 31053072 Free PMC article.
-
Metallopeptidases of Toxoplasma gondii: in silico identification and gene expression.Parasite. 2018;25:26. doi: 10.1051/parasite/2018025. Epub 2018 May 8. Parasite. 2018. PMID: 29737275 Free PMC article.
-
Post-Translational Modifications of Proteins of Malaria Parasites during the Life Cycle.Int J Mol Sci. 2024 Jun 2;25(11):6145. doi: 10.3390/ijms25116145. Int J Mol Sci. 2024. PMID: 38892332 Free PMC article. Review.
-
Metacaspases: Potential Drug Target Against Protozoan Parasites.Front Pharmacol. 2019 Jul 18;10:790. doi: 10.3389/fphar.2019.00790. eCollection 2019. Front Pharmacol. 2019. PMID: 31379569 Free PMC article. Review.
References
-
- Ranson H & Lissenden N (2016) Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol 32, 187–196. - PubMed
-
- Wells TNC, van Huijsduijnen RH & Van Voorhis WC (2015) Malaria medicines: a glass half full? Nat Rev Drug Discov 14, 424–442. - PubMed
-
- Turk B (2006) Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov 5, 785–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

