Genome analysis of the thermoacidophilic archaeon Acidianus copahuensis focusing on the metabolisms associated to biomining activities
- PMID: 28587624
- PMCID: PMC5461723
- DOI: 10.1186/s12864-017-3828-x
Genome analysis of the thermoacidophilic archaeon Acidianus copahuensis focusing on the metabolisms associated to biomining activities
Abstract
Background: Several archaeal species from the order Sulfolobales are interesting from the biotechnological point of view due to their biomining capacities. Within this group, the genus Acidianus contains four biomining species (from ten known Acidianus species), but none of these have their genome sequenced. To get insights into the genetic potential and metabolic pathways involved in the biomining activity of this group, we sequenced the genome of Acidianus copahuensis ALE1 strain, a novel thermoacidophilic crenarchaeon (optimum growth: 75 °C, pH 3) isolated from the volcanic geothermal area of Copahue at Neuquén province in Argentina. Previous experimental characterization of A. copahuensis revealed a high biomining potential, exhibited as high oxidation activity of sulfur and sulfur compounds, ferrous iron and sulfide minerals (e.g.: pyrite). This strain is also autotrophic and tolerant to heavy metals, thus, it can grow under adverse conditions for most forms of life with a low nutrient demand, conditions that are commonly found in mining environments.
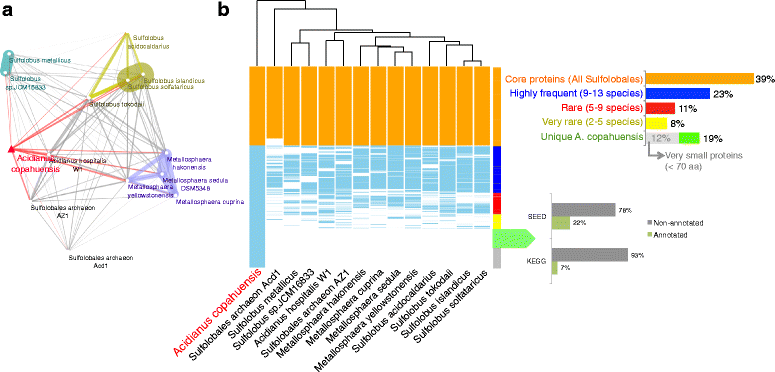
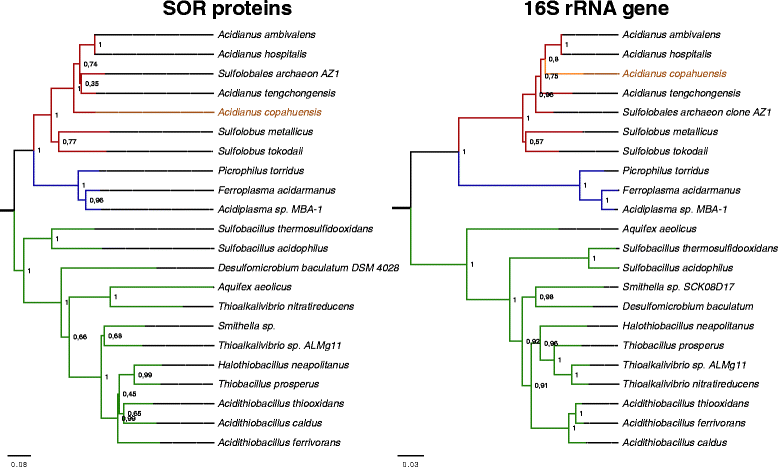
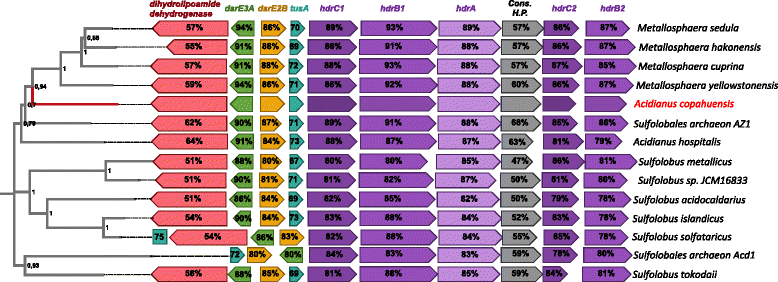
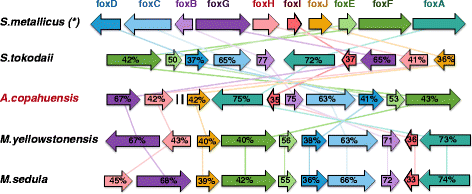
Results: In this work we analyzed the genome of Acidianus copahuensis and describe the genetic pathways involved in biomining processes. We identified the enzymes that are most likely involved in growth on sulfur and ferrous iron oxidation as well as those involved in autotrophic carbon fixation. We also found that A. copahuensis genome gathers different features that are only present in particular lineages or species from the order Sulfolobales, some of which are involved in biomining. We found that although most of its genes (81%) were found in at least one other Sulfolobales species, it is not specifically closer to any particular species (60-70% of proteins shared with each of them). Although almost one fifth of A. copahuensis proteins are not found in any other Sulfolobales species, most of them corresponded to hypothetical proteins from uncharacterized metabolisms.
Conclusion: In this work we identified the genes responsible for the biomining metabolisms that we have previously observed experimentally. We provide a landscape of the metabolic potentials of this strain in the context of Sulfolobales and propose various pathways and cellular processes not yet fully understood that can use A. copahuensis as an experimental model to further understand the fascinating biology of thermoacidophilic biomining archaea.
Keywords: Acidianus copahuensis; Biomining genes; Thermoacidophilic archaea.
Figures
Similar articles
-
Draft Genome Sequence of the Novel Thermoacidophilic Archaeon Acidianus copahuensis Strain ALE1, Isolated from the Copahue Volcanic Area in Neuquen, Argentina.Genome Announc. 2014 May 8;2(3):e00259-14. doi: 10.1128/genomeA.00259-14. Genome Announc. 2014. PMID: 24812211 Free PMC article.
-
Physiologic versatility and growth flexibility as the main characteristics of a novel thermoacidophilic Acidianus strain isolated from Copahue geothermal area in Argentina.Microb Ecol. 2013 Feb;65(2):336-46. doi: 10.1007/s00248-012-0129-4. Epub 2012 Oct 3. Microb Ecol. 2013. PMID: 23052926
-
The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism.Appl Environ Microbiol. 2008 Feb;74(3):682-92. doi: 10.1128/AEM.02019-07. Epub 2007 Dec 14. Appl Environ Microbiol. 2008. PMID: 18083856 Free PMC article.
-
Prokaryotic sulfur oxidation.Curr Opin Microbiol. 2005 Jun;8(3):253-9. doi: 10.1016/j.mib.2005.04.005. Curr Opin Microbiol. 2005. PMID: 15939347 Review.
-
Physiology, Taxonomy, and Sulfur Metabolism of the Sulfolobales, an Order of Thermoacidophilic Archaea.Front Microbiol. 2021 Oct 14;12:768283. doi: 10.3389/fmicb.2021.768283. eCollection 2021. Front Microbiol. 2021. PMID: 34721370 Free PMC article. Review.
Cited by
-
Life in hot acid: a genome-based reassessment of the archaeal order Sulfolobales.Environ Microbiol. 2021 Jul;23(7):3568-3584. doi: 10.1111/1462-2920.15189. Epub 2020 Sep 7. Environ Microbiol. 2021. PMID: 32776389 Free PMC article.
-
Roles and Regulation of Quorum Sensing of Acidophiles in Bioleaching: A Review.Microorganisms. 2024 Feb 20;12(3):422. doi: 10.3390/microorganisms12030422. Microorganisms. 2024. PMID: 38543473 Free PMC article. Review.
-
Wood-Ljungdahl pathway found in novel marine Korarchaeota groups illuminates their evolutionary history.mSystems. 2023 Aug 31;8(4):e0030523. doi: 10.1128/msystems.00305-23. Epub 2023 Jul 17. mSystems. 2023. PMID: 37458475 Free PMC article.
-
Increased chalcopyrite bioleaching capabilities of extremely thermoacidophilic Metallosphaera sedula inocula by mixotrophic propagation.J Ind Microbiol Biotechnol. 2019 Aug;46(8):1113-1127. doi: 10.1007/s10295-019-02193-3. Epub 2019 Jun 5. J Ind Microbiol Biotechnol. 2019. PMID: 31165968
-
The Proposed Molecular Mechanisms Used by Archaea for Fe(III) Reduction and Fe(II) Oxidation.Front Microbiol. 2021 Jul 1;12:690918. doi: 10.3389/fmicb.2021.690918. eCollection 2021. Front Microbiol. 2021. PMID: 34276623 Free PMC article. Review.
References
-
- Donati ER, Sand W. Microbial processing of metal sulfides. 130th vol. Dordrecht: Springer; 2007.
-
- Wang S. Copper leaching from chalcopyrite concentrates. JOM. 2005;57:48–51. doi: 10.1007/s11837-005-0252-5. - DOI
-
- Johnson DB, Okibe N, Wakeman K, Yajie L. Effect of temperature on the bioleaching of chalcopyrite concentrates containing different concentrations of silver. Hydrometallurgy. 2008;94:42–47. doi: 10.1016/j.hydromet.2008.06.005. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

