Seneca Valley Virus Suppresses Host Type I Interferon Production by Targeting Adaptor Proteins MAVS, TRIF, and TANK for Cleavage
- PMID: 28566380
- PMCID: PMC5533933
- DOI: 10.1128/JVI.00823-17
Seneca Valley Virus Suppresses Host Type I Interferon Production by Targeting Adaptor Proteins MAVS, TRIF, and TANK for Cleavage
Abstract
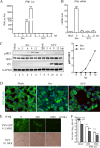
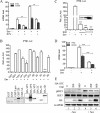
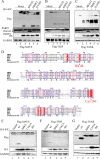
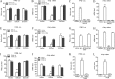
Seneca Valley virus (SVV) is an oncolytic RNA virus belonging to the Picornaviridae family. Its nucleotide sequence is highly similar to those of members of the Cardiovirus genus. SVV is also a neuroendocrine cancer-selective oncolytic picornavirus that can be used for anticancer therapy. However, the interaction between SVV and its host is yet to be fully characterized. In this study, SVV inhibited antiviral type I interferon (IFN) responses by targeting different host adaptors, including mitochondrial antiviral signaling (MAVS), Toll/interleukin 1 (IL-1) receptor domain-containing adaptor inducing IFN-β (TRIF), and TRAF family member-associated NF-κB activator (TANK), via viral 3C protease (3Cpro). SVV 3Cpro mediated the cleavage of MAVS, TRIF, and TANK at specific sites, which required its protease activity. The cleaved MAVS, TRIF, and TANK lost the ability to regulate pattern recognition receptor (PRR)-mediated IFN production. The cleavage of TANK also facilitated TRAF6-induced NF-κB activation. SVV was also found to be sensitive to IFN-β. Therefore, SVV suppressed antiviral IFN production to escape host antiviral innate immune responses by cleaving host adaptor molecules.IMPORTANCE Host cells have developed various defenses against microbial pathogen infection. The production of IFN is the first line of defense against microbial infection. However, viruses have evolved many strategies to disrupt this host defense. SVV, a member of the Picornavirus genus, is an oncolytic virus that shows potential functions in anticancer therapy. It has been demonstrated that IFN can be used in anticancer therapy for certain tumors. However, the relationship between oncolytic virus and innate immune response in anticancer therapy is still not well known. In this study, we showed that SVV has evolved as an effective mechanism to inhibit host type I IFN production by using its 3Cpro to cleave the molecules MAVS, TRIF, and TANK directly. These molecules are crucial for the Toll-like receptor 3 (TLR3)-mediated and retinoic acid-inducible gene I (RIG-I)-like receptor (RLR)-mediated signaling pathway. We also found that SVV is sensitive to IFN-β. These findings increase our understanding of the interaction between SVV and host innate immunity.
Keywords: 3C-like protease; MAVS; RNA virus; Seneca Valley virus; TANK; TRIF; cell signaling; innate immunity; interferons.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling.PLoS Pathog. 2011 Mar;7(3):e1001311. doi: 10.1371/journal.ppat.1001311. Epub 2011 Mar 10. PLoS Pathog. 2011. PMID: 21436888 Free PMC article.
-
Seneca Valley virus 2C and 3C inhibit type I interferon production by inducing the degradation of RIG-I.Virology. 2019 Sep;535:122-129. doi: 10.1016/j.virol.2019.06.017. Epub 2019 Jun 28. Virology. 2019. PMID: 31299488
-
Seneca Valley Virus 3C protease negatively regulates the type I interferon pathway by acting as a viral deubiquitinase.Antiviral Res. 2018 Dec;160:183-189. doi: 10.1016/j.antiviral.2018.10.028. Epub 2018 Nov 5. Antiviral Res. 2018. PMID: 30408499 Free PMC article.
-
How Dengue Virus Circumvents Innate Immunity.Front Immunol. 2018 Dec 4;9:2860. doi: 10.3389/fimmu.2018.02860. eCollection 2018. Front Immunol. 2018. PMID: 30564245 Free PMC article. Review.
-
Mechanisms and pathways of innate immune activation and regulation in health and cancer.Hum Vaccin Immunother. 2014;10(11):3270-85. doi: 10.4161/21645515.2014.979640. Hum Vaccin Immunother. 2014. PMID: 25625930 Free PMC article. Review.
Cited by
-
Avian Metapneumovirus Subgroup C Phosphoprotein Suppresses Type I Interferon Production by Blocking Interferon Regulatory Factor 3 Nuclear Translocation.Microbiol Spectr. 2023 Feb 14;11(1):e0341322. doi: 10.1128/spectrum.03413-22. Epub 2022 Dec 20. Microbiol Spectr. 2023. PMID: 36537793 Free PMC article.
-
Feline Infectious Peritonitis Virus Nsp5 Inhibits Type I Interferon Production by Cleaving NEMO at Multiple Sites.Viruses. 2019 Dec 30;12(1):43. doi: 10.3390/v12010043. Viruses. 2019. PMID: 31905881 Free PMC article.
-
Porcine Epidemic Diarrhea Virus and the Host Innate Immune Response.Pathogens. 2020 May 11;9(5):367. doi: 10.3390/pathogens9050367. Pathogens. 2020. PMID: 32403318 Free PMC article. Review.
-
Seneca Valley virus 3C protease cleaves OPTN (optineurin) to Impair selective autophagy and type I interferon signaling.Autophagy. 2024 Mar;20(3):614-628. doi: 10.1080/15548627.2023.2277108. Epub 2023 Nov 6. Autophagy. 2024. PMID: 37930946 Free PMC article.
-
Mouse adaptation of the H9N2 avian influenza virus causes the downregulation of genes related to innate immune responses and ubiquitin-mediated proteolysis in mice.Med Microbiol Immunol. 2020 Apr;209(2):151-161. doi: 10.1007/s00430-020-00656-4. Epub 2020 Jan 25. Med Microbiol Immunol. 2020. PMID: 31982962 Free PMC article.
References
-
- Rudin CM, Poirier JT, Senzer NN, Stephenson J, Loesch D, Burroughs KD, Reddy PS, Hann CL, Hallenbeck PL. 2011. Phase I clinical study of Seneca Valley virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res 17:888–895. doi:10.1158/1078-0432.CCR-10-1706. - DOI - PMC - PubMed
-
- Morton CL, Houghton PJ, Kolb EA, Gorlick R, Reynolds CP, Kang MH, Maris JM, Keir ST, Wu J, Smith MA. 2010. Initial testing of the replication competent Seneca Valley virus (NTX-010) by the pediatric preclinical testing program. Pediatr Blood Cancer 55:295–303. doi:10.1002/pbc.22535. - DOI - PMC - PubMed
-
- Liu Z, Zhao X, Mao H, Baxter PA, Huang Y, Yu L, Wadhwa L, Su JM, Adesina A, Perlaky L, Hurwitz M, Idamakanti N, Police SR, Hallenbeck PL, Hurwitz RL, Lau CC, Chintagumpala M, Blaney SM, Li XN. 2013. Intravenous injection of oncolytic picornavirus SVV-001 prolongs animal survival in a panel of primary tumor-based orthotopic xenograft mouse models of pediatric glioma. Neuro Oncol 15:1173–1185. doi:10.1093/neuonc/not065. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

