Viral DNA Replication Orientation and hnRNPs Regulate Transcription of the Human Papillomavirus 18 Late Promoter
- PMID: 28559488
- PMCID: PMC5449659
- DOI: 10.1128/mBio.00713-17
Viral DNA Replication Orientation and hnRNPs Regulate Transcription of the Human Papillomavirus 18 Late Promoter
Abstract
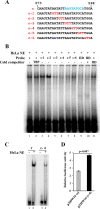
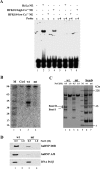
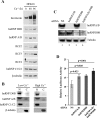
The life cycle of human papillomaviruses (HPVs) is tightly linked to keratinocyte differentiation. Although expression of viral early genes is initiated immediately upon virus infection of undifferentiated basal cells, viral DNA amplification and late gene expression occur only in the mid to upper strata of the keratinocytes undergoing terminal differentiation. In this report, we show that the relative activity of HPV18 TATA-less late promoter P811 depends on its orientation relative to that of the origin (Ori) of viral DNA replication and is sensitive to the eukaryotic DNA polymerase inhibitor aphidicolin. Additionally, transfected 70-nucleotide (nt)-long single-strand DNA oligonucleotides that are homologous to the region near Ori induce late promoter activity. We also found that promoter activation in raft cultures leads to production of the late promoter-associated, sense-strand transcription initiation RNAs (tiRNAs) and splice-site small RNAs (spliRNAs). Finally, a cis-acting AAGTATGCA core element that functions as a repressor to the promoter was identified. This element interacts with hnRNP D0B and hnRNP A/B factors. Point mutations in the core prevented binding of hnRNPs and increased the promoter activity. Confirming this result, knocking down the expression of both hnRNPs in keratinocytes led to increased promoter activity. Taking the data together, our study revealed the mechanism of how the HPV18 late promoter is regulated by DNA replication and host factors.IMPORTANCE It has been known for decades that the activity of viral late promoters is associated with viral DNA replication among almost all DNA viruses. However, the mechanism of how DNA replication activates the viral late promoter and what components of the replication machinery are involved remain largely unknown. In this study, we characterized the P811 promoter region of HPV18 and demonstrated that its activation depends on the orientation of DNA replication. Using single-stranded oligonucleotides targeting the replication fork on either leading or lagging strands, we showed that viral lagging-strand replication activates the promoter. We also identified a transcriptional repressor element located upstream of the promoter transcription start site which interacts with cellular proteins hnRNP D0B and hnRNP A/B and modulates the late promoter activity. This is the first report on how DNA replication activates a viral late promoter.
Keywords: DNA replication; HPV18; gene expression; human papillomaviruses; promoters; small RNAs; transcription; transcriptional regulation.
Copyright © 2017 Wang et al.
Figures
Similar articles
-
Keratinocyte Differentiation-Dependent Human Papillomavirus Gene Regulation.Viruses. 2017 Aug 30;9(9):245. doi: 10.3390/v9090245. Viruses. 2017. PMID: 28867768 Free PMC article. Review.
-
The Nuclear DNA Sensor IFI16 Acts as a Restriction Factor for Human Papillomavirus Replication through Epigenetic Modifications of the Viral Promoters.J Virol. 2015 Aug;89(15):7506-20. doi: 10.1128/JVI.00013-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972554 Free PMC article.
-
Serine/Arginine-Rich Splicing Factor 3 and Heterogeneous Nuclear Ribonucleoprotein A1 Regulate Alternative RNA Splicing and Gene Expression of Human Papillomavirus 18 through Two Functionally Distinguishable cis Elements.J Virol. 2016 Sep 29;90(20):9138-52. doi: 10.1128/JVI.00965-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489271 Free PMC article.
-
Brd4 Activates Early Viral Transcription upon Human Papillomavirus 18 Infection of Primary Keratinocytes.mBio. 2016 Nov 22;7(6):e01644-16. doi: 10.1128/mBio.01644-16. mBio. 2016. PMID: 27879331 Free PMC article.
-
Control of human papillomavirus gene expression by alternative splicing.Virus Res. 2017 Mar 2;231:83-95. doi: 10.1016/j.virusres.2016.11.016. Epub 2016 Nov 17. Virus Res. 2017. PMID: 27867028 Free PMC article. Review.
Cited by
-
A Genome-Wide Epstein-Barr Virus Polyadenylation Map and Its Antisense RNA to EBNA.J Virol. 2019 Jan 4;93(2):e01593-18. doi: 10.1128/JVI.01593-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30355690 Free PMC article.
-
Manipulation of Epithelial Differentiation by HPV Oncoproteins.Viruses. 2019 Apr 22;11(4):369. doi: 10.3390/v11040369. Viruses. 2019. PMID: 31013597 Free PMC article. Review.
-
Current Status of Human Papillomavirus-Related Head and Neck Cancer: From Viral Genome to Patient Care.Virol Sin. 2021 Dec;36(6):1284-1302. doi: 10.1007/s12250-021-00413-8. Epub 2021 Jun 21. Virol Sin. 2021. PMID: 34152564 Free PMC article. Review.
-
Keratinocyte Differentiation-Dependent Human Papillomavirus Gene Regulation.Viruses. 2017 Aug 30;9(9):245. doi: 10.3390/v9090245. Viruses. 2017. PMID: 28867768 Free PMC article. Review.
-
The POU-HD TFs impede the replication efficiency of several human papillomavirus genomes.Virol J. 2024 Mar 5;21(1):54. doi: 10.1186/s12985-024-02334-w. Virol J. 2024. PMID: 38444021 Free PMC article.
References
-
- Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ, International Agency for Research on Cancer Multicenter Cervical Cancer Study Group . 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348:518–527. doi:10.1056/NEJMoa021641. - DOI - PubMed
-
- Zheng ZM. 2014. Human papillomaviruses, p 87–112. In Yarchoan R. (), Cancers in people with HIV and AIDS. Springer, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

