Lineage Establishment and Progression within the Inner Cell Mass of the Mouse Blastocyst Requires FGFR1 and FGFR2
- PMID: 28552559
- PMCID: PMC5530874
- DOI: 10.1016/j.devcel.2017.05.003
Lineage Establishment and Progression within the Inner Cell Mass of the Mouse Blastocyst Requires FGFR1 and FGFR2
Abstract
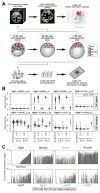
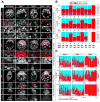
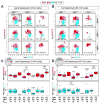
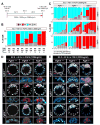
Fibroblast growth factor 4 (FGF4) is the key signal driving specification of primitive endoderm (PrE) versus pluripotent epiblast (EPI) within the inner cell mass (ICM) of the mouse blastocyst. To gain insight into the receptor(s) responding to FGF4 within ICM cells, we combined single-cell-resolution quantitative imaging with single-cell transcriptomics of wild-type and Fgf receptor (Fgfr) mutant embryos. Despite the PrE-specific expression of FGFR2, it is FGFR1, expressed by all ICM cells, that is critical for establishment of a PrE identity. Signaling through FGFR1 is also required to constrain levels of the pluripotency-associated factor NANOG in EPI cells. However, the activity of both receptors is required for lineage establishment within the ICM. Gene expression profiling of 534 single ICM cells identified distinct downstream targets associated with each receptor. These data lead us to propose a model whereby unique and additive activities of FGFR1 and FGFR2 within the ICM coordinate establishment of two distinct lineages.
Keywords: FGF; GATA6; NANOG; blastocyst; epiblast; inner cell mass; pluripotency; primitive endoderm; quantitative image analysis; single-cell transcriptomics.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Distinct Requirements for FGFR1 and FGFR2 in Primitive Endoderm Development and Exit from Pluripotency.Dev Cell. 2017 Jun 5;41(5):511-526.e4. doi: 10.1016/j.devcel.2017.05.004. Epub 2017 May 25. Dev Cell. 2017. PMID: 28552557 Free PMC article.
-
Allocation of inner cells to epiblast vs primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8→16- and 16→32-cells).Dev Biol. 2014 Jan 1;385(1):136-48. doi: 10.1016/j.ydbio.2013.09.008. Epub 2013 Sep 13. Dev Biol. 2014. PMID: 24041854
-
Distinct mechanisms for PDGF and FGF signaling in primitive endoderm development.Dev Biol. 2018 Oct 1;442(1):155-161. doi: 10.1016/j.ydbio.2018.07.010. Epub 2018 Jul 17. Dev Biol. 2018. PMID: 30026121 Free PMC article.
-
[Epiblast and primitive endoderm cell specification during mouse preimplantation development: a combination between biology and mathematical modeling].Med Sci (Paris). 2016 Feb;32(2):192-7. doi: 10.1051/medsci/20163202013. Epub 2016 Mar 2. Med Sci (Paris). 2016. PMID: 26936177 Review. French.
-
A close look at the mammalian blastocyst: epiblast and primitive endoderm formation.Cell Mol Life Sci. 2014 Sep;71(17):3327-38. doi: 10.1007/s00018-014-1630-3. Epub 2014 May 4. Cell Mol Life Sci. 2014. PMID: 24794628 Free PMC article. Review.
Cited by
-
Capturing and Interconverting Embryonic Cell Fates in a Dish.Curr Top Dev Biol. 2018;128:181-202. doi: 10.1016/bs.ctdb.2017.11.008. Epub 2018 Jan 12. Curr Top Dev Biol. 2018. PMID: 29477163 Free PMC article. Review.
-
The transition from local to global patterns governs the differentiation of mouse blastocysts.PLoS One. 2020 May 15;15(5):e0233030. doi: 10.1371/journal.pone.0233030. eCollection 2020. PLoS One. 2020. PMID: 32413083 Free PMC article.
-
Loss of KANSL3 leads to defective inner cell mass and early embryonic lethality.Mol Reprod Dev. 2024 May;91(5):e23760. doi: 10.1002/mrd.23760. Mol Reprod Dev. 2024. PMID: 38769918 Free PMC article.
-
Interleukin-6 promotes primitive endoderm development in bovine blastocysts.BMC Dev Biol. 2021 Jan 12;21(1):3. doi: 10.1186/s12861-020-00235-z. BMC Dev Biol. 2021. PMID: 33430761 Free PMC article.
-
Super-enhancers conserved within placental mammals maintain stem cell pluripotency.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2204716119. doi: 10.1073/pnas.2204716119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161929 Free PMC article.
References
-
- Behringer R, Gertsenstein M, Nagy KV, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. 4. Cold Spring Harbor Laboratory Press; Cold Spring harbor, NY: 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

