Distinct Requirements for FGFR1 and FGFR2 in Primitive Endoderm Development and Exit from Pluripotency
- PMID: 28552557
- PMCID: PMC5502766
- DOI: 10.1016/j.devcel.2017.05.004
Distinct Requirements for FGFR1 and FGFR2 in Primitive Endoderm Development and Exit from Pluripotency
Abstract
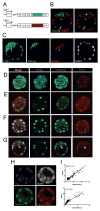
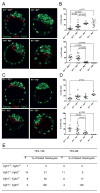
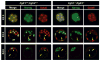
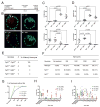
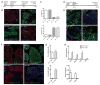
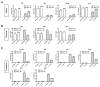
Activation of the FGF signaling pathway during preimplantation development of the mouse embryo is known to be essential for differentiation of the inner cell mass and the formation of the primitive endoderm (PrE). We now show using fluorescent reporter knockin lines that Fgfr1 is expressed in all cell populations of the blastocyst, while Fgfr2 expression becomes restricted to extraembryonic lineages, including the PrE. We further show that loss of both receptors prevents the development of the PrE and demonstrate that FGFR1 plays a more prominent role in this process than FGFR2. Finally, we document an essential role for FGFRs in embryonic stem cell (ESC) differentiation, with FGFR1 again having a greater influence than FGFR2 in ESC exit from the pluripotent state. Collectively, these results identify mechanisms through which FGF signaling regulates inner cell mass lineage restriction and cell commitment during preimplantation development.
Keywords: FGF; epiblast; pluripotency; primitive endoderm; signaling.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Lineage Establishment and Progression within the Inner Cell Mass of the Mouse Blastocyst Requires FGFR1 and FGFR2.Dev Cell. 2017 Jun 5;41(5):496-510.e5. doi: 10.1016/j.devcel.2017.05.003. Epub 2017 May 25. Dev Cell. 2017. PMID: 28552559 Free PMC article.
-
Distinct mechanisms for PDGF and FGF signaling in primitive endoderm development.Dev Biol. 2018 Oct 1;442(1):155-161. doi: 10.1016/j.ydbio.2018.07.010. Epub 2018 Jul 17. Dev Biol. 2018. PMID: 30026121 Free PMC article.
-
Allocation of inner cells to epiblast vs primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8→16- and 16→32-cells).Dev Biol. 2014 Jan 1;385(1):136-48. doi: 10.1016/j.ydbio.2013.09.008. Epub 2013 Sep 13. Dev Biol. 2014. PMID: 24041854
-
Lineage specification in the early mouse embryo.Exp Cell Res. 2014 Feb 1;321(1):32-9. doi: 10.1016/j.yexcr.2013.12.004. Epub 2013 Dec 11. Exp Cell Res. 2014. PMID: 24333597 Review.
-
A close look at the mammalian blastocyst: epiblast and primitive endoderm formation.Cell Mol Life Sci. 2014 Sep;71(17):3327-38. doi: 10.1007/s00018-014-1630-3. Epub 2014 May 4. Cell Mol Life Sci. 2014. PMID: 24794628 Free PMC article. Review.
Cited by
-
Growth-factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development.Elife. 2020 Jul 28;9:e56079. doi: 10.7554/eLife.56079. Elife. 2020. PMID: 32720894 Free PMC article.
-
Origin and function of the yolk sac in primate embryogenesis.Nat Commun. 2020 Jul 28;11(1):3760. doi: 10.1038/s41467-020-17575-w. Nat Commun. 2020. PMID: 32724077 Free PMC article. Review.
-
FGFR1 regulates trophectoderm development and facilitates blastocyst implantation.Dev Biol. 2019 Feb 1;446(1):94-101. doi: 10.1016/j.ydbio.2018.12.008. Epub 2018 Dec 12. Dev Biol. 2019. PMID: 30552867 Free PMC article.
-
Suppression of ERK signalling abolishes primitive endoderm formation but does not promote pluripotency in rabbit embryo.Development. 2017 Oct 15;144(20):3719-3730. doi: 10.1242/dev.156406. Epub 2017 Sep 21. Development. 2017. PMID: 28935706 Free PMC article.
-
A NANOG-pERK reciprocal regulatory circuit regulates Nanog autoregulation and ERK signaling dynamics.EMBO Rep. 2022 Nov 7;23(11):e54421. doi: 10.15252/embr.202154421. Epub 2022 Sep 6. EMBO Rep. 2022. PMID: 36066347 Free PMC article.
References
-
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5082–5087. - PMC - PubMed
-
- Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, Dupont G, Chazaud C. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development (Cambridge, England) 2014;141:3637–3648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

