Efficacy of β-lactam/β-lactamase inhibitor combination is linked to WhiB4-mediated changes in redox physiology of Mycobacterium tuberculosis
- PMID: 28548640
- PMCID: PMC5473688
- DOI: 10.7554/eLife.25624
Efficacy of β-lactam/β-lactamase inhibitor combination is linked to WhiB4-mediated changes in redox physiology of Mycobacterium tuberculosis
Abstract
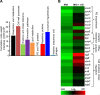
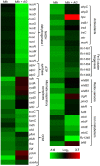
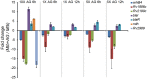
Mycobacterium tuberculosis (Mtb) expresses a broad-spectrum β-lactamase (BlaC) that mediates resistance to one of the highly effective antibacterials, β-lactams. Nonetheless, β-lactams showed mycobactericidal activity in combination with β-lactamase inhibitor, clavulanate (Clav). However, the mechanistic aspects of how Mtb responds to β-lactams such as Amoxicillin in combination with Clav (referred as Augmentin [AG]) are not clear. Here, we identified cytoplasmic redox potential and intracellular redox sensor, WhiB4, as key determinants of mycobacterial resistance against AG. Using computer-based, biochemical, redox-biosensor, and genetic strategies, we uncovered a functional linkage between specific determinants of β-lactam resistance (e.g. β-lactamase) and redox potential in Mtb. We also describe the role of WhiB4 in coordinating the activity of β-lactamase in a redox-dependent manner to tolerate AG. Disruption of WhiB4 enhances AG tolerance, whereas overexpression potentiates AG activity against drug-resistant Mtb. Our findings suggest that AG can be exploited to diminish drug-resistance in Mtb through redox-based interventions.
Keywords: Amoxicillin-Clavulanate; Mycobacterium tuberculosis; Mycothiol; Peptidoglycan; drug tolerance; infectious disease; microbiology; persistence.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




Similar articles
-
Meropenem-clavulanate has high in vitro activity against multidrug-resistant Mycobacterium tuberculosis.Int J Mycobacteriol. 2015 Mar;4 Suppl 1:80-1. doi: 10.1016/j.ijmyco.2014.10.018. Epub 2014 Nov 11. Int J Mycobacteriol. 2015. PMID: 27128620
-
Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis β-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of β-lactam-β-lactamase inhibitor combinations.Antimicrob Agents Chemother. 2013 Dec;57(12):6085-96. doi: 10.1128/AAC.01253-13. Epub 2013 Sep 23. Antimicrob Agents Chemother. 2013. PMID: 24060876 Free PMC article.
-
In Vitro Activity of β-Lactams in Combination with β-Lactamase Inhibitors against Mycobacterium tuberculosis Clinical Isolates.Biomed Res Int. 2018 Jul 2;2018:3579832. doi: 10.1155/2018/3579832. eCollection 2018. Biomed Res Int. 2018. PMID: 30065936 Free PMC article.
-
Have we realized the full potential of β-lactams for treating drug-resistant TB?IUBMB Life. 2018 Sep;70(9):881-888. doi: 10.1002/iub.1875. Epub 2018 Jun 22. IUBMB Life. 2018. PMID: 29934998 Free PMC article. Review.
-
The In vitro activity of carbapenems alone and in combination with β-lactamase inhibitors against difficult-to-treat mycobacteria; Mycobacterium tuberculosis, Mycobacterium abscessus, and Mycobacterium avium complex: A systematic review.Int J Mycobacteriol. 2023 Jul-Sep;12(3):211-225. doi: 10.4103/ijmy.ijmy_131_23. Int J Mycobacteriol. 2023. PMID: 37721224 Review.
Cited by
-
Targeting redox heterogeneity to counteract drug tolerance in replicating Mycobacterium tuberculosis.Sci Transl Med. 2019 Nov 13;11(518):eaaw6635. doi: 10.1126/scitranslmed.aaw6635. Sci Transl Med. 2019. PMID: 31723039 Free PMC article.
-
A path-based analysis of infected cell line and COVID-19 patient transcriptome reveals novel potential targets and drugs against SARS-CoV-2.Res Sq [Preprint]. 2022 Mar 21:rs.3.rs-1474136. doi: 10.21203/rs.3.rs-1474136/v1. Res Sq. 2022. Update in: Front Immunol. 2022 Jul 01;13:918817. doi: 10.3389/fimmu.2022.918817. PMID: 35434729 Free PMC article. Updated. Preprint.
-
Stable integration of the Mrx1-roGFP2 biosensor to monitor dynamic changes of the mycothiol redox potential in Corynebacterium glutamicum.Redox Biol. 2019 Jan;20:514-525. doi: 10.1016/j.redox.2018.11.012. Epub 2018 Nov 17. Redox Biol. 2019. PMID: 30481728 Free PMC article.
-
Selection of Multi-Drug Targets against Drug-Resistant Mycobacterium tuberculosis XDR1219 Using the Hyperbolic Mapping of the Protein Interaction Network.Int J Mol Sci. 2023 Sep 13;24(18):14050. doi: 10.3390/ijms241814050. Int J Mol Sci. 2023. PMID: 37762354 Free PMC article.
-
Genome-Wide Transcriptional Responses of Mycobacterium to Antibiotics.Front Microbiol. 2019 Feb 20;10:249. doi: 10.3389/fmicb.2019.00249. eCollection 2019. Front Microbiol. 2019. PMID: 30842759 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

