Systems-wide Studies Uncover Commander, a Multiprotein Complex Essential to Human Development
- PMID: 28544880
- PMCID: PMC5541947
- DOI: 10.1016/j.cels.2017.04.006
Systems-wide Studies Uncover Commander, a Multiprotein Complex Essential to Human Development
Abstract
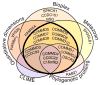
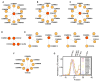
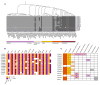
Recent mass spectrometry maps of the human interactome independently support the existence of a large multiprotein complex, dubbed "Commander." Broadly conserved across animals and ubiquitously expressed in nearly every human cell type examined thus far, Commander likely plays a fundamental cellular function, akin to other ubiquitous machines involved in expression, proteostasis, and trafficking. Experiments on individual subunits support roles in endosomal protein sorting, including the trafficking of Notch proteins, copper transporters, and lipoprotein receptors. Commander is critical for vertebrate embryogenesis, and defects in the complex and its interaction partners disrupt craniofacial, brain, and heart development. Here, we review the synergy between large-scale proteomic efforts and focused studies in the discovery of Commander, describe its composition, structure, and function, and discuss how it illustrates the power of systems biology. Based on 3D modeling and biochemical data, we draw strong parallels between Commander and the retromer cargo-recognition complex, laying a foundation for future research into Commander's role in human developmental disorders.
Keywords: Commander complex; developmental disorders; endosomal protein sorting; mammalian interactome; multiprotein complex; system-wide studies.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Retromer and sorting nexins in endosomal sorting.Biochem Soc Trans. 2015 Feb;43(1):33-47. doi: 10.1042/BST20140290. Biochem Soc Trans. 2015. PMID: 25619244 Review.
-
Commander Complex-A Multifaceted Operator in Intracellular Signaling and Cargo.Cells. 2021 Dec 7;10(12):3447. doi: 10.3390/cells10123447. Cells. 2021. PMID: 34943955 Free PMC article. Review.
-
Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling.Nat Cell Biol. 2017 Oct;19(10):1214-1225. doi: 10.1038/ncb3610. Epub 2017 Sep 11. Nat Cell Biol. 2017. PMID: 28892079 Free PMC article.
-
Updated Insight into the Physiological and Pathological Roles of the Retromer Complex.Int J Mol Sci. 2017 Jul 25;18(8):1601. doi: 10.3390/ijms18081601. Int J Mol Sci. 2017. PMID: 28757549 Free PMC article. Review.
-
Structure and interactions of the endogenous human Commander complex.Nat Struct Mol Biol. 2024 Jun;31(6):925-938. doi: 10.1038/s41594-024-01246-1. Epub 2024 Mar 8. Nat Struct Mol Biol. 2024. PMID: 38459129 Free PMC article.
Cited by
-
Noncanonical roles of ATG5 and membrane atg8ylation in retromer assembly and function.bioRxiv [Preprint]. 2024 Oct 14:2024.07.10.602886. doi: 10.1101/2024.07.10.602886. bioRxiv. 2024. PMID: 39026874 Free PMC article. Preprint.
-
Does Subtelomeric Position of COMMD5 Influence Cancer Progression?Front Oncol. 2021 Mar 9;11:642130. doi: 10.3389/fonc.2021.642130. eCollection 2021. Front Oncol. 2021. PMID: 33768002 Free PMC article.
-
Structural basis for Retriever-SNX17 assembly and endosomal sorting.bioRxiv [Preprint]. 2024 Mar 13:2024.03.12.584676. doi: 10.1101/2024.03.12.584676. bioRxiv. 2024. Update in: Nat Commun. 2024 Nov 25;15(1):10193. doi: 10.1038/s41467-024-54583-6 PMID: 38559023 Free PMC article. Updated. Preprint.
-
Ancient eukaryotic protein interactions illuminate modern genetic traits and disorders.bioRxiv [Preprint]. 2024 May 29:2024.05.26.595818. doi: 10.1101/2024.05.26.595818. bioRxiv. 2024. PMID: 38853926 Free PMC article. Preprint.
-
Receptor Recycling by Retromer.Mol Cell Biol. 2023;43(7):317-334. doi: 10.1080/10985549.2023.2222053. Epub 2023 Jun 23. Mol Cell Biol. 2023. PMID: 37350516 Free PMC article. Review.
References
-
- Bartuzi P, Hofker MH, van de Sluis B. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochimica et Biophysica Acta. 2013;1832:2315–2321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

