Analysis of early mesothelial cell responses to Staphylococcus epidermidis isolated from patients with peritoneal dialysis-associated peritonitis
- PMID: 28542390
- PMCID: PMC5443531
- DOI: 10.1371/journal.pone.0178151
Analysis of early mesothelial cell responses to Staphylococcus epidermidis isolated from patients with peritoneal dialysis-associated peritonitis
Abstract
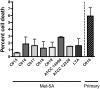
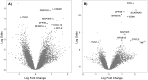
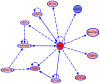
The major complication of peritoneal dialysis (PD) is the development of peritonitis, an infection within the abdominal cavity, primarily caused by bacteria. PD peritonitis is associated with significant morbidity, mortality and health care costs. Staphylococcus epidermidis is the most frequently isolated cause of PD-associated peritonitis. Mesothelial cells are integral to the host response to peritonitis, and subsequent clinical outcomes, yet the effects of infection on mesothelial cells are not well characterised. We systematically investigated the early mesothelial cell response to clinical and reference isolates of S. epidermidis using primary mesothelial cells and the mesothelial cell line Met-5A. Using an unbiased whole genome microarray, followed by a targeted panel of genes known to be involved in the human antibacterial response, we identified 38 differentially regulated genes (adj. p-value < 0.05) representing 35 canonical pathways after 1 hour exposure to S. epidermidis. The top 3 canonical pathways were TNFR2 signaling, IL-17A signaling, and TNFR1 signaling (adj. p-values of 0.0012, 0.0012 and 0.0019, respectively). Subsequent qPCR validation confirmed significant differences in gene expression in a number of genes not previously described in mesothelial cell responses to infection, with heterogeneity observed between clinical isolates of S. epidermidis, and between Met-5A and primary mesothelial cells. Heterogeneity between different S. epidermidis isolates suggests that specific virulence factors may play critical roles in influencing outcomes from peritonitis. This study provides new insights into early mesothelial cell responses to infection with S. epidermidis, and confirms the importance of validating findings in primary mesothelial cells.
Conflict of interest statement
Figures

Similar articles
-
Low immunogenicity allows Staphylococcus epidermidis to cause PD peritonitis.Perit Dial Int. 2011 Nov-Dec;31(6):672-8. doi: 10.3747/pdi.2009.00150. Epub 2010 May 6. Perit Dial Int. 2011. PMID: 20448241
-
Establishing an experimental infection model for peritoneal dialysis: effect of inoculum and volume.Perit Dial Int. 1993;13 Suppl 2:S79-80. Perit Dial Int. 1993. PMID: 8399677
-
Challenging the current treatment paradigm for methicillin-resistant Staphylococcus epidermidis peritonitis in peritoneal dialysis patients.Perit Dial Int. 2002 May-Jun;22(3):335-8. Perit Dial Int. 2002. PMID: 12227390
-
Peritonitis: finding success by evaluating the failures.Int J Artif Organs. 1996 Dec;19(12):695-6. Int J Artif Organs. 1996. PMID: 9029243 Review. No abstract available.
-
Pathophysiological changes to the peritoneal membrane during PD-related peritonitis: the role of mesothelial cells.Mediators Inflamm. 2012;2012:484167. doi: 10.1155/2012/484167. Epub 2012 Apr 10. Mediators Inflamm. 2012. PMID: 22577250 Free PMC article. Review.
Cited by
-
Analysis of the Ribonuclease A Superfamily of Antimicrobial Peptides in Patients Undergoing Chronic Peritoneal Dialysis.Sci Rep. 2019 May 23;9(1):7753. doi: 10.1038/s41598-019-44219-x. Sci Rep. 2019. PMID: 31123272 Free PMC article.
-
Fracture biomechanics influence local and systemic immune responses in a murine fracture-related infection model.Biol Open. 2021 Sep 15;10(9):bio057315. doi: 10.1242/bio.057315. Epub 2021 Oct 1. Biol Open. 2021. PMID: 34240122 Free PMC article.
-
Managing abdominal cocoon syndrome complicated by intestinal necrosis and unexpected amelioration of depression after surgery: a case report.J Med Case Rep. 2024 Jul 6;18(1):322. doi: 10.1186/s13256-024-04650-9. J Med Case Rep. 2024. PMID: 38970114 Free PMC article.
References
-
- Cass A, Chadban S, Gallagher M, Howard K, Jones A, McDonald S, et al. The economic impact of End-Stage Kidney Disease in Australia: Projections to 2020. Kidney Health Australia [Internet]. 2010.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

