TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton
- PMID: 28537271
- PMCID: PMC5458083
- DOI: 10.1038/ncomms15588
TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton
Abstract
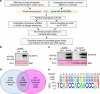
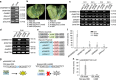
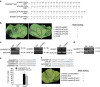
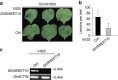
Transcription activator-like (TAL) effectors from Xanthomonas citri subsp. malvacearum (Xcm) are essential for bacterial blight of cotton (BBC). Here, by combining transcriptome profiling with TAL effector-binding element (EBE) prediction, we show that GhSWEET10, encoding a functional sucrose transporter, is induced by Avrb6, a TAL effector determining Xcm pathogenicity. Activation of GhSWEET10 by designer TAL effectors (dTALEs) restores virulence of Xcm avrb6 deletion strains, whereas silencing of GhSWEET10 compromises cotton susceptibility to infections. A BBC-resistant line carrying an unknown recessive b6 gene bears the same EBE as the susceptible line, but Avrb6-mediated induction of GhSWEET10 is reduced, suggesting a unique mechanism underlying b6-mediated resistance. We show via an extensive survey of GhSWEET transcriptional responsiveness to different Xcm field isolates that additional GhSWEETs may also be involved in BBC. These findings advance our understanding of the disease and resistance in cotton and may facilitate the development cotton with improved resistance to BBC.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Identification of a virulence tal gene in the cotton pathogen, Xanthomonas citri pv. malvacearum strain Xss-V2-18.BMC Microbiol. 2020 Apr 15;20(1):91. doi: 10.1186/s12866-020-01783-x. BMC Microbiol. 2020. PMID: 32293266 Free PMC article.
-
Two TAL Effectors of Xanthomonas citri pv. malvacearum Induce Water Soaking by Activating GhSWEET14 Genes in Cotton.Mol Plant Pathol. 2025 Jan;26(1):e70053. doi: 10.1111/mpp.70053. Mol Plant Pathol. 2025. PMID: 39825471 Free PMC article.
-
Activation of three targets by a TAL effector confers susceptibility to bacterial blight of cotton.Nat Commun. 2025 Jan 14;16(1):644. doi: 10.1038/s41467-025-55926-7. Nat Commun. 2025. PMID: 39809734 Free PMC article.
-
Plant Executor Genes.Int J Mol Sci. 2022 Jan 28;23(3):1524. doi: 10.3390/ijms23031524. Int J Mol Sci. 2022. PMID: 35163443 Free PMC article. Review.
-
Xanthomonas AvrBs3 family-type III effectors: discovery and function.Annu Rev Phytopathol. 2010;48:419-36. doi: 10.1146/annurev-phyto-080508-081936. Annu Rev Phytopathol. 2010. PMID: 19400638 Review.
Cited by
-
Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants.Plant Mol Biol. 2019 Jul;100(4-5):351-365. doi: 10.1007/s11103-019-00872-4. Epub 2019 Apr 27. Plant Mol Biol. 2019. PMID: 31030374 Review.
-
Starving the enemy: how plant and microbe compete for sugar on the border.Front Plant Sci. 2023 Aug 2;14:1230254. doi: 10.3389/fpls.2023.1230254. eCollection 2023. Front Plant Sci. 2023. PMID: 37600180 Free PMC article. Review.
-
Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease.Mol Plant Pathol. 2018 Sep;19(9):2149-2161. doi: 10.1111/mpp.12689. Epub 2018 Jul 19. Mol Plant Pathol. 2018. PMID: 29660235 Free PMC article.
-
Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors.BMC Genomics. 2017 Aug 30;18(1):670. doi: 10.1186/s12864-017-4087-6. BMC Genomics. 2017. PMID: 28854875 Free PMC article.
-
Nematode-resistance loci in upland cotton genomes are associated with structural differences.G3 (Bethesda). 2024 Sep 4;14(9):jkae140. doi: 10.1093/g3journal/jkae140. G3 (Bethesda). 2024. PMID: 38934790 Free PMC article.
References
-
- Paterson A. H. et al.. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012). - PubMed
-
- Wang K. et al.. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103 (2012). - PubMed
-
- Li F. et al.. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572 (2014). - PubMed
-
- Li F. et al.. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

