Structure of a pre-catalytic spliceosome
- PMID: 28530653
- PMCID: PMC5503131
- DOI: 10.1038/nature22799
Structure of a pre-catalytic spliceosome
Abstract
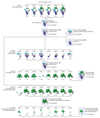
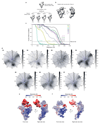
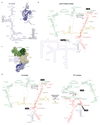
Intron removal requires assembly of the spliceosome on precursor mRNA (pre-mRNA) and extensive remodelling to form the spliceosome's catalytic centre. Here we report the cryo-electron microscopy structure of the yeast Saccharomyces cerevisiae pre-catalytic B complex spliceosome at near-atomic resolution. The mobile U2 small nuclear ribonucleoprotein particle (snRNP) associates with U4/U6.U5 tri-snRNP through the U2/U6 helix II and an interface between U4/U6 di-snRNP and the U2 snRNP SF3b-containing domain, which also transiently contacts the helicase Brr2. The 3' region of the U2 snRNP is flexibly attached to the SF3b-containing domain and protrudes over the concave surface of tri-snRNP, where the U1 snRNP may reside before its release from the pre-mRNA 5' splice site. The U6 ACAGAGA sequence forms a hairpin that weakly tethers the 5' splice site. The B complex proteins Prp38, Snu23 and Spp381 bind the Prp8 N-terminal domain and stabilize U6 ACAGAGA stem-pre-mRNA and Brr2-U4 small nuclear RNA interactions. These results provide important insights into the events leading to active site formation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





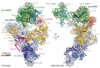
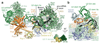


Similar articles
-
Cryo-EM structure of the spliceosome immediately after branching.Nature. 2016 Sep 8;537(7619):197-201. doi: 10.1038/nature19316. Epub 2016 Jul 26. Nature. 2016. PMID: 27459055 Free PMC article.
-
Prespliceosome structure provides insights into spliceosome assembly and regulation.Nature. 2018 Jul;559(7714):419-422. doi: 10.1038/s41586-018-0323-8. Epub 2018 Jul 11. Nature. 2018. PMID: 29995849 Free PMC article.
-
Structure of a spliceosome remodelled for exon ligation.Nature. 2017 Feb 16;542(7641):377-380. doi: 10.1038/nature21078. Epub 2017 Jan 11. Nature. 2017. PMID: 28076345 Free PMC article.
-
How Is Precursor Messenger RNA Spliced by the Spliceosome?Annu Rev Biochem. 2020 Jun 20;89:333-358. doi: 10.1146/annurev-biochem-013118-111024. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815536 Review.
-
RNA Splicing by the Spliceosome.Annu Rev Biochem. 2020 Jun 20;89:359-388. doi: 10.1146/annurev-biochem-091719-064225. Epub 2019 Dec 3. Annu Rev Biochem. 2020. PMID: 31794245 Review.
Cited by
-
Single-Molecule Analysis of Reverse Transcriptase Enzymes.Cold Spring Harb Perspect Biol. 2019 Sep 3;11(9):a032458. doi: 10.1101/cshperspect.a032458. Cold Spring Harb Perspect Biol. 2019. PMID: 31481455 Free PMC article. Review.
-
Frozen in time: analyzing molecular dynamics with time-resolved cryo-EM.Structure. 2023 Jan 5;31(1):4-19. doi: 10.1016/j.str.2022.11.014. Epub 2022 Dec 29. Structure. 2023. PMID: 36584678 Free PMC article. Review.
-
Recruitment of a splicing factor to the nuclear lamina for its inactivation.Commun Biol. 2022 Jul 22;5(1):736. doi: 10.1038/s42003-022-03689-y. Commun Biol. 2022. PMID: 35869234 Free PMC article.
-
The SMN complex drives structural changes in human snRNAs to enable snRNP assembly.Nat Commun. 2023 Oct 18;14(1):6580. doi: 10.1038/s41467-023-42324-0. Nat Commun. 2023. PMID: 37852981 Free PMC article.
-
A brave new world of RNA-binding proteins.Nat Rev Mol Cell Biol. 2018 May;19(5):327-341. doi: 10.1038/nrm.2017.130. Epub 2018 Jan 17. Nat Rev Mol Cell Biol. 2018. PMID: 29339797 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

