RNA Pol II Dynamics Modulate Co-transcriptional Chromatin Modification, CTD Phosphorylation, and Transcriptional Direction
- PMID: 28506463
- PMCID: PMC5488731
- DOI: 10.1016/j.molcel.2017.04.016
RNA Pol II Dynamics Modulate Co-transcriptional Chromatin Modification, CTD Phosphorylation, and Transcriptional Direction
Abstract
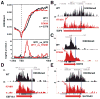
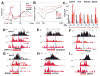
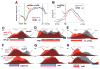
Eukaryotic genes are marked by conserved post-translational modifications on the RNA pol II C-terminal domain (CTD) and the chromatin template. How the 5'-3' profiles of these marks are established is poorly understood. Using pol II mutants in human cells, we found that slow transcription repositioned specific co-transcriptionally deposited chromatin modifications; histone H3 lysine 36 trimethyl (H3K36me3) shifted within genes toward 5' ends, and histone H3 lysine 4 dimethyl (H3K4me2) extended farther upstream of start sites. Slow transcription also evoked a hyperphosphorylation of CTD Ser2 residues at 5' ends of genes that is conserved in yeast. We propose a "dwell time in the target zone" model to explain the effects of transcriptional dynamics on the establishment of co-transcriptionally deposited protein modifications. Promoter-proximal Ser2 phosphorylation is associated with a longer pol II dwell time at start sites and reduced transcriptional polarity because of strongly enhanced divergent antisense transcription at promoters. These results demonstrate that pol II dynamics help govern the decision between sense and divergent antisense transcription.
Keywords: H3K4me2; K3K36me3; antisense transcription; bidirectional transcription; histone methylation; kinetic coupling; pol II CTD S2 phosphorylation; pol II dynamics; transcription elongation rate.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Gene-specific RNA polymerase II phosphorylation and the CTD code.Nat Struct Mol Biol. 2010 Oct;17(10):1279-86. doi: 10.1038/nsmb.1913. Epub 2010 Sep 12. Nat Struct Mol Biol. 2010. PMID: 20835241 Free PMC article.
-
Phospho-site mutants of the RNA Polymerase II C-terminal domain alter subtelomeric gene expression and chromatin modification state in fission yeast.Nucleic Acids Res. 2016 Nov 2;44(19):9180-9189. doi: 10.1093/nar/gkw603. Epub 2016 Jul 8. Nucleic Acids Res. 2016. PMID: 27402158 Free PMC article.
-
A feed forward circuit comprising Spt6, Ctk1 and PAF regulates Pol II CTD phosphorylation and transcription elongation.Nucleic Acids Res. 2014 Jan;42(2):870-81. doi: 10.1093/nar/gkt1003. Epub 2013 Oct 25. Nucleic Acids Res. 2014. PMID: 24163256 Free PMC article.
-
Gene-specific requirement of RNA polymerase II CTD phosphorylation.Mol Microbiol. 2012 Jun;84(6):995-1004. doi: 10.1111/j.1365-2958.2012.08071.x. Epub 2012 May 4. Mol Microbiol. 2012. PMID: 22553990 Review.
-
Transcription by RNA polymerase II and the CTD-chromatin crosstalk.Biochem Biophys Res Commun. 2022 Apr 9;599:81-86. doi: 10.1016/j.bbrc.2022.02.039. Epub 2022 Feb 11. Biochem Biophys Res Commun. 2022. PMID: 35176629 Review.
Cited by
-
MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation.Mol Cell. 2019 May 16;74(4):674-687.e11. doi: 10.1016/j.molcel.2019.02.031. Epub 2019 Mar 27. Mol Cell. 2019. PMID: 30928206 Free PMC article.
-
Open chromatin analysis in Trypanosoma cruzi life forms highlights critical differences in genomic compartments and developmental regulation at tDNA loci.Epigenetics Chromatin. 2022 Jun 1;15(1):22. doi: 10.1186/s13072-022-00450-x. Epigenetics Chromatin. 2022. PMID: 35650626 Free PMC article.
-
Distinct roles of two SEC scaffold proteins, AFF1 and AFF4, in regulating RNA polymerase II transcription elongation.J Mol Cell Biol. 2024 Jan 17;15(8):mjad049. doi: 10.1093/jmcb/mjad049. J Mol Cell Biol. 2024. PMID: 37528066 Free PMC article.
-
Identification of a novel base J binding protein complex involved in RNA polymerase II transcription termination in trypanosomes.PLoS Genet. 2020 Feb 21;16(2):e1008390. doi: 10.1371/journal.pgen.1008390. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32084124 Free PMC article.
-
Truncating the spliceosomal 'rope protein' Prp45 results in Htz1 dependent phenotypes.RNA Biol. 2024 Jan;21(1):1-17. doi: 10.1080/15476286.2024.2348896. Epub 2024 May 6. RNA Biol. 2024. PMID: 38711165 Free PMC article.
References
-
- Aebi M, Weissman C. Precision and orderliness in splicing. Trends in Genetics. 1987;3:102–107.
-
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

