Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18
- PMID: 28506460
- PMCID: PMC5523955
- DOI: 10.1016/j.molcel.2017.04.009
Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18
Abstract
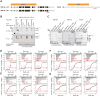
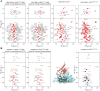
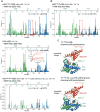
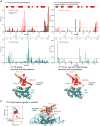
The protein 53BP1 plays a central regulatory role in DNA double-strand break repair. 53BP1 relocates to chromatin by recognizing RNF168-mediated mono-ubiquitylation of histone H2A Lys15 in the nucleosome core particle dimethylated at histone H4 Lys20 (NCP-ubme). 53BP1 relocation is terminated by ubiquitin ligases RNF169 and RAD18 via unknown mechanisms. Using nuclear magnetic resonance (NMR) spectroscopy and biochemistry, we show that RNF169 bridges ubiquitin and histone surfaces, stabilizing a pre-existing ubiquitin orientation in NCP-ubme to form a high-affinity complex. This conformational selection mechanism contrasts with the low-affinity binding mode of 53BP1, and it ensures 53BP1 displacement by RNF169 from NCP-ubme. We also show that RAD18 binds tightly to NCP-ubme through a ubiquitin-binding domain that contacts ubiquitin and nucleosome surfaces accessed by 53BP1. Our work uncovers diverse ubiquitin recognition mechanisms in the nucleosome, explaining how RNF168, RNF169, and RAD18 regulate 53BP1 chromatin recruitment and how specificity can be achieved in the recognition of a ubiquitin-modified substrate.
Keywords: 53BP1; DNA repair; NMR spectroscopy; RAD18; RNF168; RNF169; biophysics; nucleosome; structural biology; ubiquitylation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
The structural basis of modified nucleosome recognition by 53BP1.Nature. 2016 Aug 4;536(7614):100-3. doi: 10.1038/nature18951. Epub 2016 Jul 27. Nature. 2016. PMID: 27462807
-
Mechanisms of RNF168 nucleosome recognition and ubiquitylation.Mol Cell. 2024 Mar 7;84(5):839-853.e12. doi: 10.1016/j.molcel.2023.12.036. Epub 2024 Jan 18. Mol Cell. 2024. PMID: 38242129 Free PMC article.
-
Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks.J Cell Biol. 2012 Apr 16;197(2):189-99. doi: 10.1083/jcb.201109100. Epub 2012 Apr 9. J Cell Biol. 2012. PMID: 22492721 Free PMC article.
-
New answers to the old RIDDLE: RNF168 and the DNA damage response pathway.FEBS J. 2022 May;289(9):2467-2480. doi: 10.1111/febs.15857. Epub 2021 Apr 16. FEBS J. 2022. PMID: 33797206 Free PMC article. Review.
-
Regulatory ubiquitylation in response to DNA double-strand breaks.DNA Repair (Amst). 2009 Apr 5;8(4):436-43. doi: 10.1016/j.dnarep.2009.01.013. Epub 2009 Feb 18. DNA Repair (Amst). 2009. PMID: 19230794 Review.
Cited by
-
53BP1: Keeping It under Control, Even at a Distance from DNA Damage.Genes (Basel). 2022 Dec 16;13(12):2390. doi: 10.3390/genes13122390. Genes (Basel). 2022. PMID: 36553657 Free PMC article. Review.
-
53BP1 Enforces Distinct Pre- and Post-resection Blocks on Homologous Recombination.Mol Cell. 2020 Jan 2;77(1):26-38.e7. doi: 10.1016/j.molcel.2019.09.024. Epub 2019 Oct 22. Mol Cell. 2020. PMID: 31653568 Free PMC article.
-
Ectopic RNF168 expression promotes break-induced replication-like DNA synthesis at stalled replication forks.Nucleic Acids Res. 2020 May 7;48(8):4298-4308. doi: 10.1093/nar/gkaa154. Nucleic Acids Res. 2020. PMID: 32182354 Free PMC article.
-
Defective repair of topoisomerase I induced chromosomal damage in Huntington's disease.Cell Mol Life Sci. 2022 Feb 28;79(3):160. doi: 10.1007/s00018-022-04204-6. Cell Mol Life Sci. 2022. PMID: 35224690 Free PMC article.
-
Ubiquitylation in DNA double-strand break repair.DNA Repair (Amst). 2021 Jul;103:103129. doi: 10.1016/j.dnarep.2021.103129. Epub 2021 May 7. DNA Repair (Amst). 2021. PMID: 33990032 Free PMC article.
References
-
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. - PubMed
-
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

