Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth
- PMID: 28500171
- PMCID: PMC5542851
- DOI: 10.1182/blood-2016-11-751099
Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth
Abstract
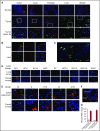
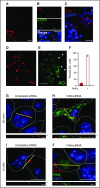
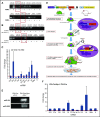
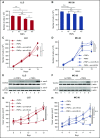
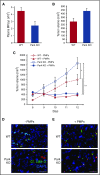
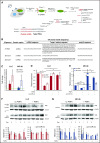
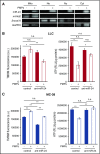
Platelet-derived microparticles (PMPs) are associated with enhancement of metastasis and poor cancer outcomes. Circulating PMPs transfer platelet microRNAs (miRNAs) to vascular cells. Solid tumor vasculature is highly permeable, allowing the possibility of PMP-tumor cell interaction. Here, we show that PMPs infiltrate solid tumors in humans and mice and transfer platelet-derived RNA, including miRNAs, to tumor cells in vivo and in vitro, resulting in tumor cell apoptosis. MiR-24 was a major species in this transfer. PMP transfusion inhibited growth of both lung and colon carcinoma ectopic tumors, whereas blockade of miR-24 in tumor cells accelerated tumor growth in vivo, and prevented tumor growth inhibition by PMPs. Conversely, Par4-deleted mice, which had reduced circulating microparticles (MPs), supported accelerated tumor growth which was halted by PMP transfusion. PMP targeting was associated with tumor cell apoptosis in vivo. We identified direct RNA targets of platelet-derived miR-24 in tumor cells, which included mitochondrial mt-Nd2, and Snora75, a noncoding small nucleolar RNA. These RNAs were suppressed in PMP-treated tumor cells, resulting in mitochondrial dysfunction and growth inhibition, in an miR-24-dependent manner. Thus, platelet-derived miRNAs transfer in vivo to tumor cells in solid tumors via infiltrating MPs, regulate tumor cell gene expression, and modulate tumor progression. These findings provide novel insight into mechanisms of horizontal RNA transfer and add multiple layers to the regulatory roles of miRNAs and PMPs in tumor progression. Plasma MP-mediated transfer of regulatory RNAs and modulation of gene expression may be a common feature with important outcomes in contexts of enhanced vascular permeability.
© 2017 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
Comment in
-
Platelet-derived nanomedicine targets cancer.Blood. 2017 Aug 3;130(5):561-562. doi: 10.1182/blood-2017-05-786327. Blood. 2017. PMID: 28775157 No abstract available.
-
Platelet microparticles: small payloads with profound effects on tumor growth.Noncoding RNA Investig. 2017 Nov;1:15. doi: 10.21037/ncri.2017.11.02. Epub 2017 Nov 17. Noncoding RNA Investig. 2017. PMID: 31218277 Free PMC article. No abstract available.
Similar articles
-
Platelet Microparticles and miRNA Transfer in Cancer Progression: Many Targets, Modes of Action, and Effects Across Cancer Stages.Front Cardiovasc Med. 2018 Feb 28;5:13. doi: 10.3389/fcvm.2018.00013. eCollection 2018. Front Cardiovasc Med. 2018. PMID: 29564336 Free PMC article.
-
Platelet microparticle delivered microRNA-Let-7a promotes the angiogenic switch.Biochim Biophys Acta Mol Basis Dis. 2018 Aug;1864(8):2633-2643. doi: 10.1016/j.bbadis.2018.04.013. Epub 2018 Apr 21. Biochim Biophys Acta Mol Basis Dis. 2018. PMID: 29684582
-
Synergistic effects of EMPs and PMPs on pulmonary vascular leakage and lung injury after ischemia/reperfusion.Cell Commun Signal. 2020 Nov 23;18(1):184. doi: 10.1186/s12964-020-00672-0. Cell Commun Signal. 2020. PMID: 33225929 Free PMC article.
-
Role of platelet-derived microparticles in angiogenesis and tumor progression.Discov Med. 2009 Dec;8(43):237-41. Discov Med. 2009. PMID: 20040277 Review.
-
Platelet microparticles and cancer: An intimate cross-talk.Transfus Apher Sci. 2015 Oct;53(2):168-72. doi: 10.1016/j.transci.2015.10.014. Epub 2015 Oct 27. Transfus Apher Sci. 2015. PMID: 26542350 Review.
Cited by
-
Platelet Microparticle Controversial Role in Cancer.Adv Pharm Bull. 2021 Jan;11(1):39-55. doi: 10.34172/apb.2021.005. Epub 2020 Nov 7. Adv Pharm Bull. 2021. PMID: 33747851 Free PMC article. Review.
-
Tumor-Educated Platelet RNA for the Detection and (Pseudo)progression Monitoring of Glioblastoma.Cell Rep Med. 2020 Oct 20;1(7):100101. doi: 10.1016/j.xcrm.2020.100101. Cell Rep Med. 2020. PMID: 33103128 Free PMC article.
-
Platelet count as a biomarker for monitoring treatment response and disease recurrence in recurrent epithelial ovarian cancer.J Ovarian Res. 2020 Jul 18;13(1):78. doi: 10.1186/s13048-020-00682-z. J Ovarian Res. 2020. PMID: 32682445 Free PMC article.
-
The role of microvesicles and its active molecules in regulating cellular biology.J Cell Mol Med. 2019 Dec;23(12):7894-7904. doi: 10.1111/jcmm.14667. Epub 2019 Sep 27. J Cell Mol Med. 2019. PMID: 31559684 Free PMC article. Review.
-
Microparticles: biogenesis, characteristics and intervention therapy for cancers in preclinical and clinical research.J Nanobiotechnology. 2022 Apr 13;20(1):189. doi: 10.1186/s12951-022-01358-0. J Nanobiotechnology. 2022. PMID: 35418077 Free PMC article. Review.
References
-
- Camerer E, Qazi AA, Duong DN, Cornelissen I, Advincula R, Coughlin SR. Platelets, protease-activated receptors, and fibrinogen in hematogenous metastasis. Blood. 2004;104(2):397-401. - PubMed
-
- Gay LJ, Felding-Habermann B. Platelets alter tumor cell attributes to propel metastasis: programming in transit. Cancer Cell. 2011;20(5):553-554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

