Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava
- PMID: 28498327
- PMCID: PMC5454953
- DOI: 10.3390/ijms18051041
Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava
Abstract
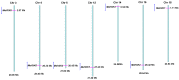
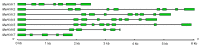
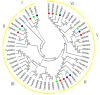
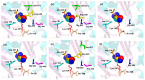
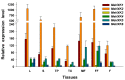
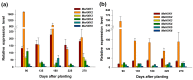
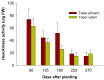
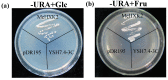
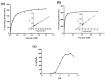
Hexokinase (HXK) proteins play important roles in catalyzing hexose phosphorylation and sugar sensing and signaling. To investigate the roles of HXKs in cassava tuber root development, seven HXK genes (MeHXK1-7) were isolated and analyzed. A phylogenetic analysis revealed that the MeHXK family can be divided into five subfamilies of plant HXKs. MeHXKs were clearly divided into type A (MeHXK1) and type B (MeHXK2-7) based on their N-terminal sequences. MeHXK1-5 all had typical conserved regions and similar protein structures to the HXKs of other plants; while MeHXK6-7 lacked some of the conserved regions. An expression analysis of the MeHXK genes in cassava organs or tissues demonstrated that MeHXK2 is the dominant HXK in all the examined tissues (leaves, stems, fruits, tuber phloems, and tuber xylems). Notably, the expression of MeHXK2 and the enzymatic activity of HXK were higher at the initial and expanding tuber stages, and lower at the mature tuber stage. Furthermore, the HXK activity of MeHXK2 was identified by functional complementation of the HXK-deficient yeast strain YSH7.4-3C (hxk1, hxk2, glk1). The gene expression and enzymatic activity of MeHXK2 suggest that it might be the main enzyme for hexose phosphorylation during cassava tuber root development, which is involved in sucrose metabolism to regulate the accumulation of starch.
Keywords: cassava; enzyme activities; gene expression; hexokinase; yeast complementation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



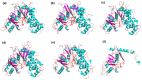
Similar articles
-
Identification, Expression, and Functional Analysis of the Fructokinase Gene Family in Cassava.Int J Mol Sci. 2017 Nov 12;18(11):2398. doi: 10.3390/ijms18112398. Int J Mol Sci. 2017. PMID: 29137155 Free PMC article.
-
The involvement of hexokinase in the coordinated regulation of glucose and gibberellin on cell wall invertase and sucrose synthesis in grape berry.Mol Biol Rep. 2014 Dec;41(12):7899-910. doi: 10.1007/s11033-014-3683-7. Epub 2014 Aug 28. Mol Biol Rep. 2014. PMID: 25163631
-
Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.).Planta. 2006 Aug;224(3):598-611. doi: 10.1007/s00425-006-0251-y. Epub 2006 Mar 22. Planta. 2006. PMID: 16552590
-
Plant Hexokinases are Multifaceted Proteins.Plant Cell Physiol. 2017 Jul 1;58(7):1151-1160. doi: 10.1093/pcp/pcx062. Plant Cell Physiol. 2017. PMID: 28449056 Review.
-
Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development.J Exp Bot. 2014 Mar;65(3):809-19. doi: 10.1093/jxb/ert400. Epub 2013 Nov 30. J Exp Bot. 2014. PMID: 24293612 Review.
Cited by
-
Transcriptome Analysis on Key Metabolic Pathways in Rhodotorula mucilaginosa Under Pb(II) Stress.Appl Environ Microbiol. 2022 Apr 12;88(7):e0221521. doi: 10.1128/aem.02215-21. Epub 2022 Mar 21. Appl Environ Microbiol. 2022. PMID: 35311507 Free PMC article.
-
The hexokinase Gene Family in Cotton: Genome-Wide Characterization and Bioinformatics Analysis.Front Plant Sci. 2022 May 16;13:882587. doi: 10.3389/fpls.2022.882587. eCollection 2022. Front Plant Sci. 2022. PMID: 35651774 Free PMC article.
-
MeABL5, an ABA Insensitive 5-Like Basic Leucine Zipper Transcription Factor, Positively Regulates MeCWINV3 in Cassava (Manihot esculenta Crantz).Front Plant Sci. 2019 Jun 28;10:772. doi: 10.3389/fpls.2019.00772. eCollection 2019. Front Plant Sci. 2019. PMID: 31316528 Free PMC article.
-
Changes in sucrose metabolism patterns affect the early maturation of Cassava sexual tetraploid roots.BMC Plant Biol. 2022 Dec 10;22(1):574. doi: 10.1186/s12870-022-03969-z. BMC Plant Biol. 2022. PMID: 36496357 Free PMC article.
-
Genome-wide identification of hexokinase gene family in Brassica napus: structure, phylogenetic analysis, expression, and functional characterization.Planta. 2018 Jul;248(1):171-182. doi: 10.1007/s00425-018-2888-8. Epub 2018 Apr 11. Planta. 2018. PMID: 29644447
References
-
- Siemens J., GonzáLez M.C., Wolf S., Hofmann C., Greiner S., Du Y., Rausch T., Roitsch T., Ludwig-MüLler J. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011;12:247–262. doi: 10.1111/j.1364-3703.2010.00667.x. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

