shinyheatmap: Ultra fast low memory heatmap web interface for big data genomics
- PMID: 28493881
- PMCID: PMC5426587
- DOI: 10.1371/journal.pone.0176334
shinyheatmap: Ultra fast low memory heatmap web interface for big data genomics
Abstract
Background: Transcriptomics, metabolomics, metagenomics, and other various next-generation sequencing (-omics) fields are known for their production of large datasets, especially across single-cell sequencing studies. Visualizing such big data has posed technical challenges in biology, both in terms of available computational resources as well as programming acumen. Since heatmaps are used to depict high-dimensional numerical data as a colored grid of cells, efficiency and speed have often proven to be critical considerations in the process of successfully converting data into graphics. For example, rendering interactive heatmaps from large input datasets (e.g., 100k+ rows) has been computationally infeasible on both desktop computers and web browsers. In addition to memory requirements, programming skills and knowledge have frequently been barriers-to-entry for creating highly customizable heatmaps.
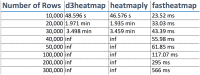
Results: We propose shinyheatmap: an advanced user-friendly heatmap software suite capable of efficiently creating highly customizable static and interactive biological heatmaps in a web browser. shinyheatmap is a low memory footprint program, making it particularly well-suited for the interactive visualization of extremely large datasets that cannot typically be computed in-memory due to size restrictions. Also, shinyheatmap features a built-in high performance web plug-in, fastheatmap, for rapidly plotting interactive heatmaps of datasets as large as 105-107 rows within seconds, effectively shattering previous performance benchmarks of heatmap rendering speed.
Conclusions: shinyheatmap is hosted online as a freely available web server with an intuitive graphical user interface: http://shinyheatmap.com. The methods are implemented in R, and are available as part of the shinyheatmap project at: https://github.com/Bohdan-Khomtchouk/shinyheatmap. Users can access fastheatmap directly from within the shinyheatmap web interface, and all source code has been made publicly available on Github: https://github.com/Bohdan-Khomtchouk/fastheatmap.
Conflict of interest statement
Figures
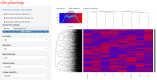
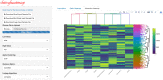
Similar articles
-
jHeatmap: an interactive heatmap viewer for the web.Bioinformatics. 2014 Jun 15;30(12):1757-8. doi: 10.1093/bioinformatics/btu094. Epub 2014 Feb 23. Bioinformatics. 2014. PMID: 24567544
-
MicroScope: ChIP-seq and RNA-seq software analysis suite for gene expression heatmaps.BMC Bioinformatics. 2016 Sep 22;17(1):390. doi: 10.1186/s12859-016-1260-x. BMC Bioinformatics. 2016. PMID: 27659774 Free PMC article.
-
HeatmapGenerator: high performance RNAseq and microarray visualization software suite to examine differential gene expression levels using an R and C++ hybrid computational pipeline.Source Code Biol Med. 2014 Dec 24;9(1):30. doi: 10.1186/s13029-014-0030-2. eCollection 2014. Source Code Biol Med. 2014. PMID: 25550709 Free PMC article.
-
ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap.Nucleic Acids Res. 2015 Jul 1;43(W1):W566-70. doi: 10.1093/nar/gkv468. Epub 2015 May 12. Nucleic Acids Res. 2015. PMID: 25969447 Free PMC article.
-
Phantasus, a web application for visual and interactive gene expression analysis.Elife. 2024 Jun 3;13:e85722. doi: 10.7554/eLife.85722. Elife. 2024. PMID: 38742735 Free PMC article.
Cited by
-
Supercritical fluid chromatography-mass spectrometry enables simultaneous measurement of all phosphoinositide regioisomers.Commun Chem. 2022 May 11;5(1):61. doi: 10.1038/s42004-022-00676-6. Commun Chem. 2022. PMID: 36697617 Free PMC article.
-
A Snapshot of the Trehalose Pathway During Seed Imbibition in Medicago truncatula Reveals Temporal- and Stress-Dependent Shifts in Gene Expression Patterns Associated With Metabolite Changes.Front Plant Sci. 2019 Dec 18;10:1590. doi: 10.3389/fpls.2019.01590. eCollection 2019. Front Plant Sci. 2019. PMID: 31921241 Free PMC article.
-
anexVis: visual analytics framework for analysis of RNA expression.Bioinformatics. 2018 Jul 15;34(14):2510-2512. doi: 10.1093/bioinformatics/bty122. Bioinformatics. 2018. PMID: 29506198 Free PMC article.
-
Unraveling the functional genes present in rhizosphere microbiomes of Solanum lycopersicum.PeerJ. 2023 Jun 2;11:e15432. doi: 10.7717/peerj.15432. eCollection 2023. PeerJ. 2023. PMID: 37283894 Free PMC article.
-
Characteristics of MicroRNA Expression Depending on the Presence or Absence of Meningioma in Patients with Neurofibromatosis Type 2: A Secondary Analysis.Neurol Med Chir (Tokyo). 2024 Mar 15;64(3):116-122. doi: 10.2176/jns-nmc.2023-0200. Epub 2024 Jan 24. Neurol Med Chir (Tokyo). 2024. PMID: 38267057 Free PMC article.
References
-
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al.: TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003, 34(2): 374–378. - PubMed
-
- Qlucore Omics Explorer: The D.I.Y Bioinformatics Software. http://www.qlucore.com.
-
- Gould J: GENE-E software hosted at the Broad Institute. http://www.broadinstitute.org/cancer/software/GENE-E/.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

