Comparative analysis of the predicted secretomes of Rosaceae scab pathogens Venturia inaequalis and V. pirina reveals expanded effector families and putative determinants of host range
- PMID: 28464870
- PMCID: PMC5412055
- DOI: 10.1186/s12864-017-3699-1
Comparative analysis of the predicted secretomes of Rosaceae scab pathogens Venturia inaequalis and V. pirina reveals expanded effector families and putative determinants of host range
Abstract
Background: Fungal plant pathogens belonging to the genus Venturia cause damaging scab diseases of members of the Rosaceae. In terms of economic impact, the most important of these are V. inaequalis, which infects apple, and V. pirina, which is a pathogen of European pear. Given that Venturia fungi colonise the sub-cuticular space without penetrating plant cells, it is assumed that effectors that contribute to virulence and determination of host range will be secreted into this plant-pathogen interface. Thus the predicted secretomes of a range of isolates of Venturia with distinct host-ranges were interrogated to reveal putative proteins involved in virulence and pathogenicity.
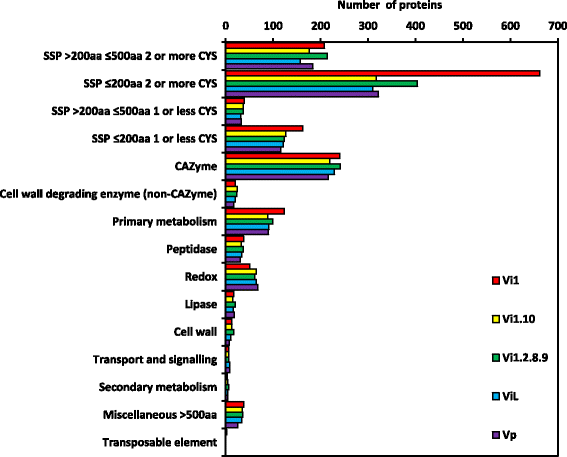
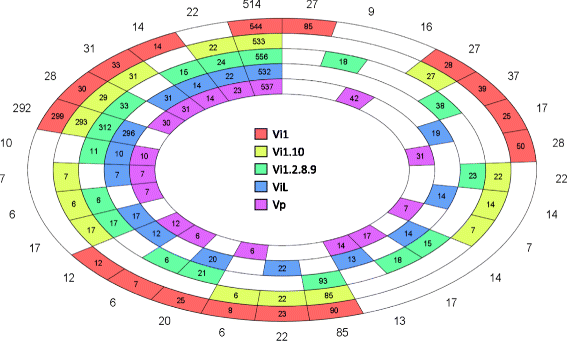
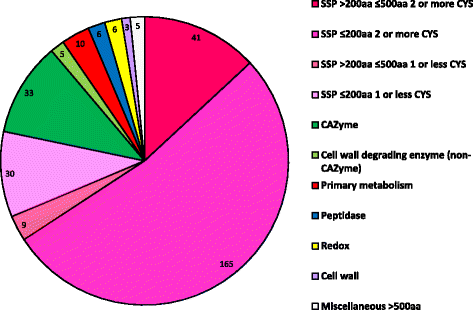
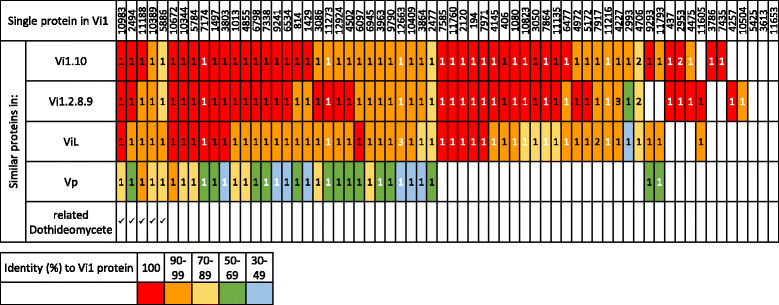
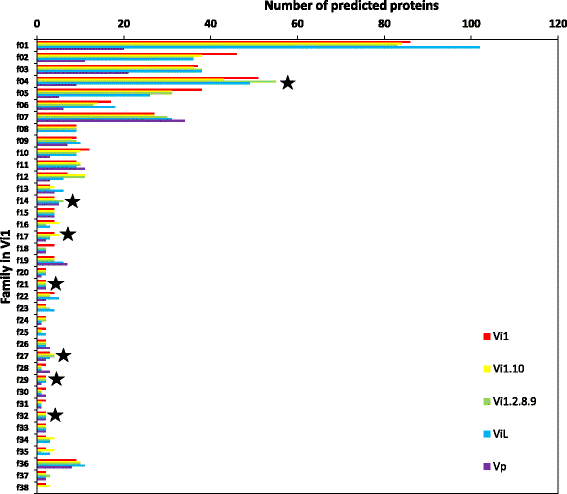
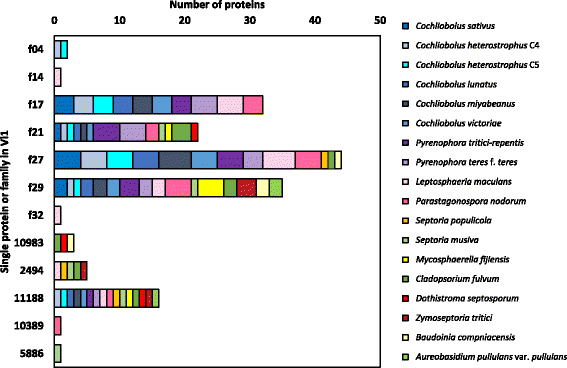
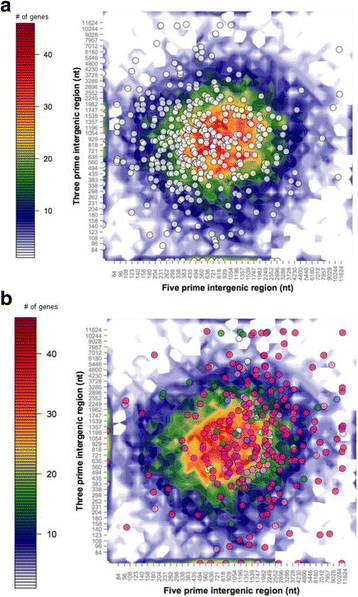
Results: Genomes of Venturia pirina (one European pear scab isolate) and Venturia inaequalis (three apple scab, and one loquat scab, isolates) were sequenced and the predicted secretomes of each isolate identified. RNA-Seq was conducted on the apple-specific V. inaequalis isolate Vi1 (in vitro and infected apple leaves) to highlight virulence and pathogenicity components of the secretome. Genes encoding over 600 small secreted proteins (candidate effectors) were identified, most of which are novel to Venturia, with expansion of putative effector families a feature of the genus. Numerous genes with similarity to Leptosphaeria maculans AvrLm6 and the Verticillium spp. Ave1 were identified. Candidates for avirulence effectors with cognate resistance genes involved in race-cultivar specificity were identified, as were putative proteins involved in host-species determination. Candidate effectors were found, on average, to be in regions of relatively low gene-density and in closer proximity to repeats (e.g. transposable elements), compared with core eukaryotic genes.
Conclusions: Comparative secretomics has revealed candidate effectors from Venturia fungal plant pathogens that attack pome fruit. Effectors that are putative determinants of host range were identified; both those that may be involved in race-cultivar and host-species specificity. Since many of the effector candidates are in close proximity to repetitive sequences this may point to a possible mechanism for the effector gene family expansion observed and a route to diversification via transposition and repeat-induced point mutation.
Keywords: Apple; Effector; European pear; Malus x domestica; Pyrus communis; Secretome; Venturia inaequalis; Venturia pirina.
Figures
Similar articles
-
A Large Family of AvrLm6-like Genes in the Apple and Pear Scab Pathogens, Venturia inaequalis and Venturia pirina.Front Plant Sci. 2015 Nov 17;6:980. doi: 10.3389/fpls.2015.00980. eCollection 2015. Front Plant Sci. 2015. PMID: 26635823 Free PMC article.
-
The Venturia inaequalis effector repertoire is dominated by expanded families with predicted structural similarity, but unrelated sequence, to avirulence proteins from other plant-pathogenic fungi.BMC Biol. 2022 Nov 3;20(1):246. doi: 10.1186/s12915-022-01442-9. BMC Biol. 2022. PMID: 36329441 Free PMC article.
-
Venturia inaequalis: the causal agent of apple scab.Mol Plant Pathol. 2011 Feb;12(2):105-22. doi: 10.1111/j.1364-3703.2010.00656.x. Epub 2010 Aug 26. Mol Plant Pathol. 2011. PMID: 21199562 Free PMC article.
-
Biology and Epidemiology of Venturia Species Affecting Fruit Crops: A Review.Front Plant Sci. 2017 Sep 19;8:1496. doi: 10.3389/fpls.2017.01496. eCollection 2017. Front Plant Sci. 2017. PMID: 28974954 Free PMC article. Review.
-
The Venturia apple pathosystem: pathogenicity mechanisms and plant defense responses.J Biomed Biotechnol. 2009;2009:680160. doi: 10.1155/2009/680160. Epub 2010 Jan 28. J Biomed Biotechnol. 2009. PMID: 20150969 Free PMC article. Review.
Cited by
-
Population Genome Sequencing of the Scab Fungal Species Venturia inaequalis, Venturia pirina, Venturia aucupariae and Venturia asperata.G3 (Bethesda). 2019 Aug 8;9(8):2405-2414. doi: 10.1534/g3.119.400047. G3 (Bethesda). 2019. PMID: 31253647 Free PMC article.
-
In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4.Biomolecules. 2021 Sep 12;11(9):1353. doi: 10.3390/biom11091353. Biomolecules. 2021. PMID: 34572566 Free PMC article.
-
Remote homology clustering identifies lowly conserved families of effector proteins in plant-pathogenic fungi.Microb Genom. 2021 Sep;7(9):000637. doi: 10.1099/mgen.0.000637. Microb Genom. 2021. PMID: 34468307 Free PMC article.
-
Cell Wall Carbohydrate Dynamics during the Differentiation of Infection Structures by the Apple Scab Fungus, Venturia inaequalis.Microbiol Spectr. 2023 Jun 15;11(3):e0421922. doi: 10.1128/spectrum.04219-22. Epub 2023 Apr 11. Microbiol Spectr. 2023. PMID: 37039647 Free PMC article.
-
A ribonuclease T2 protein FocRnt2 contributes to the virulence of Fusarium oxysporum f. sp. cubense tropical race 4.Mol Plant Pathol. 2024 Aug;25(8):e13502. doi: 10.1111/mpp.13502. Mol Plant Pathol. 2024. PMID: 39118198 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

