Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system
- PMID: 28446709
- PMCID: PMC5413908
- DOI: 10.1098/rsob.170007
Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system
Abstract
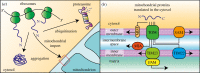
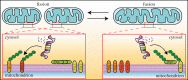
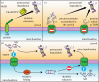
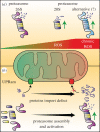
Mitochondria are pivotal organelles in eukaryotic cells. The complex proteome of mitochondria comprises proteins that are encoded by nuclear and mitochondrial genomes. The biogenesis of mitochondrial proteins requires their transport in an unfolded state with a high risk of misfolding. The mislocalization of mitochondrial proteins is deleterious to the cell. The electron transport chain in mitochondria is a source of reactive oxygen species that damage proteins. Mitochondrial dysfunction is linked to many pathological conditions and, together with the loss of cellular protein homeostasis (proteostasis), are hallmarks of ageing and ageing-related degeneration diseases. The pathogenesis of neurodegenerative disorders, such as Alzheimer's disease and Parkinson's disease, has been associated with mitochondrial and proteostasis failure. Thus, mitochondrial proteins require sophisticated surveillance mechanisms. Although mitochondria form a proteasome-exclusive compartment, multiple lines of evidence indicate a crucial role for the cytosolic ubiquitin-proteasome system (UPS) in the quality control of mitochondrial proteins. The proteasome affects mitochondrial proteins at stages of their biogenesis and maturity. The effects of the UPS go beyond the removal of damaged proteins and include the adjustment of mitochondrial proteome composition, the regulation of organelle dynamics and the protection of cellular homeostasis against mitochondrial failure. In turn, mitochondrial activity and mitochondrial dysfunction adjust the activity of the UPS, with implications at the cellular level.
Keywords: mitochondria; proteasome; protein biogenesis; proteostasis; ubiquitin; ubiquitin–proteasome system.
© 2017 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures
Similar articles
-
Mitochondrial and Ubiquitin Proteasome System Dysfunction in Ageing and Disease: Two Sides of the Same Coin?Int J Mol Sci. 2015 Aug 17;16(8):19458-76. doi: 10.3390/ijms160819458. Int J Mol Sci. 2015. PMID: 26287188 Free PMC article. Review.
-
Inhibition of proteasome reveals basal mitochondrial ubiquitination.J Proteomics. 2020 Oct 30;229:103949. doi: 10.1016/j.jprot.2020.103949. Epub 2020 Aug 31. J Proteomics. 2020. PMID: 32882436
-
Mitochondrial quality control by the ubiquitin-proteasome system.Biochem Soc Trans. 2011 Oct;39(5):1509-13. doi: 10.1042/BST0391509. Biochem Soc Trans. 2011. PMID: 21936843 Review.
-
Ubiquitin-proteasome system and mitochondria - reciprocity.Biochim Biophys Acta. 2011 Feb;1809(2):80-7. doi: 10.1016/j.bbagrm.2010.07.005. Epub 2010 Jul 30. Biochim Biophys Acta. 2011. PMID: 20674813 Review.
-
Ubiquitin-dependent mitochondrial protein degradation.Int J Biochem Cell Biol. 2011 Oct;43(10):1422-6. doi: 10.1016/j.biocel.2011.06.002. Epub 2011 Jun 12. Int J Biochem Cell Biol. 2011. PMID: 21683801 Free PMC article. Review.
Cited by
-
Serine 39 in the GTP-binding domain of Drp1 is involved in shaping mitochondrial morphology.FEBS Open Bio. 2024 Jul;14(7):1147-1165. doi: 10.1002/2211-5463.13820. Epub 2024 May 17. FEBS Open Bio. 2024. PMID: 38760979 Free PMC article.
-
The phospholipids cardiolipin and phosphatidylethanolamine differentially regulate MDC biogenesis.J Cell Biol. 2024 May 6;223(5):e202302069. doi: 10.1083/jcb.202302069. Epub 2024 Mar 18. J Cell Biol. 2024. PMID: 38497895 Free PMC article.
-
Mitophagy regulates mitochondrial number following pharmacological induction of mitochondrial biogenesis in renal proximal tubule cells.Front Pharmacol. 2024 Feb 5;15:1344075. doi: 10.3389/fphar.2024.1344075. eCollection 2024. Front Pharmacol. 2024. PMID: 38375036 Free PMC article.
-
Association of a Specific OsCULLIN3c Haplotype with Salt Stress Responses in Local Thai Rice.Int J Mol Sci. 2024 Jan 15;25(2):1040. doi: 10.3390/ijms25021040. Int J Mol Sci. 2024. PMID: 38256116 Free PMC article.
-
The roles of ubiquitination and deubiquitination of NLRP3 inflammasome in inflammation-related diseases: A review.Biomol Biomed. 2024 Jan 8;24(4):708-721. doi: 10.17305/bb.2023.9997. Biomol Biomed. 2024. PMID: 38193803 Free PMC article. Review.
References
-
- Newmeyer DD, Ferguson-Miller S. 2003. Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112, 481–490. (doi:10.1016/S0092-8674(03)00116-8) - DOI - PubMed
-
- Nunnari J, Suomalainen A. 2012. Mitochondria: in sickness and in health. Cell 148, 1145–1159. (doi:10.1016/j.cell.2012.02.035) - DOI - PMC - PubMed
-
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C. 2012. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 13, 566–578. (doi:10.1038/nrm3412) - DOI - PubMed
-
- Stehling O, Lill R. 2013. The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 5, a011312 (doi:10.1101/cshperspect.a011312) - DOI - PMC - PubMed
-
- Friedman JR, Nunnari J. 2014. Mitochondrial form and function. Nature 505, 335–343. (doi:10.1038/nature12985) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

