Comparative Genomics of Chrysochromulina Ericina Virus and Other Microalga-Infecting Large DNA Viruses Highlights Their Intricate Evolutionary Relationship with the Established Mimiviridae Family
- PMID: 28446675
- PMCID: PMC5487555
- DOI: 10.1128/JVI.00230-17
Comparative Genomics of Chrysochromulina Ericina Virus and Other Microalga-Infecting Large DNA Viruses Highlights Their Intricate Evolutionary Relationship with the Established Mimiviridae Family
Abstract
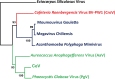
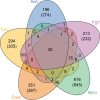
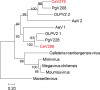
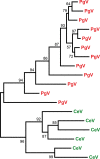
Chrysochromulina ericina virus CeV-01B (CeV) was isolated from Norwegian coastal waters in 1998. Its icosahedral particle is 160 nm in diameter and encloses a 474-kb double-stranded DNA (dsDNA) genome. This virus, although infecting a microalga (the haptophyceae Haptolina ericina, formerly Chrysochromulina ericina), is phylogenetically related to members of the Mimiviridae family, initially established with the acanthamoeba-infecting mimivirus and megavirus as prototypes. This family was later split into two genera (Mimivirus and Cafeteriavirus) following the characterization of a virus infecting the heterotrophic stramenopile Cafeteria roenbergensis (CroV). CeV, as well as two of its close relatives, which infect the unicellular photosynthetic eukaryotes Phaeocystis globosa (Phaeocystis globosa virus [PgV]) and Aureococcus anophagefferens (Aureococcus anophagefferens virus [AaV]), are currently unclassified by the International Committee on Viral Taxonomy (ICTV). The detailed comparative analysis of the CeV genome presented here confirms the phylogenetic affinity of this emerging group of microalga-infecting viruses with the Mimiviridae but argues in favor of their classification inside a distinct clade within the family. Although CeV, PgV, and AaV share more common features among them than with the larger Mimiviridae, they also exhibit a large complement of unique genes, attesting to their complex evolutionary history. We identified several gene fusion events and cases of convergent evolution involving independent lateral gene acquisitions. Finally, CeV possesses an unusual number of inteins, some of which are closely related despite being inserted in nonhomologous genes. This appears to contradict the paradigm of allele-specific inteins and suggests that the Mimiviridae are especially efficient in spreading inteins while enlarging their repertoire of homing genes.IMPORTANCE Although it infects the microalga Chrysochromulina ericina, CeV is more closely related to acanthamoeba-infecting viruses of the Mimiviridae family than to any member of the Phycodnaviridae, the ICTV-approved family historically including all alga-infecting large dsDNA viruses. CeV, as well as its relatives that infect the microalgae Phaeocystic globosa (PgV) and Aureococcus anophagefferens (AaV), remains officially unclassified and a source of confusion in the literature. Our comparative analysis of the CeV genome in the context of this emerging group of alga-infecting viruses suggests that they belong to a distinct clade within the established Mimiviridae family. The presence of a large number of unique genes as well as specific gene fusion events, evolutionary convergences, and inteins integrated at unusual locations document the complex evolutionary history of the CeV lineage.
Keywords: Aureococcus anophagefferens virus; Chrysochromulina ericina virus; Haptolina ericina virus; Megamimivirinae; Mesomimivirinae; Mimiviridae; Phaeocystis globosa virus; nucleocytoplasmic virus.
Copyright © 2017 American Society for Microbiology.
Figures





Similar articles
-
The 474-Kilobase-Pair Complete Genome Sequence of CeV-01B, a Virus Infecting Haptolina (Chrysochromulina) ericina (Prymnesiophyceae).Genome Announc. 2015 Dec 3;3(6):e01413-15. doi: 10.1128/genomeA.01413-15. Genome Announc. 2015. PMID: 26634761 Free PMC article.
-
Phylogenetic analysis of members of the Phycodnaviridae virus family, using amplified fragments of the major capsid protein gene.Appl Environ Microbiol. 2008 May;74(10):3048-57. doi: 10.1128/AEM.02548-07. Epub 2008 Mar 21. Appl Environ Microbiol. 2008. PMID: 18359826 Free PMC article.
-
Isolation and Identification of a Large Green Alga Virus (Chlorella Virus XW01) of Mimiviridae and Its Virophage (Chlorella Virus Virophage SW01) by Using Unicellular Green Algal Cultures.J Virol. 2022 Apr 13;96(7):e0211421. doi: 10.1128/jvi.02114-21. Epub 2022 Mar 9. J Virol. 2022. PMID: 35262372 Free PMC article.
-
Mimiviridae: An Expanding Family of Highly Diverse Large dsDNA Viruses Infecting a Wide Phylogenetic Range of Aquatic Eukaryotes.Viruses. 2018 Sep 18;10(9):506. doi: 10.3390/v10090506. Viruses. 2018. PMID: 30231528 Free PMC article. Review.
-
Mimivirus and its virophage.Annu Rev Genet. 2009;43:49-66. doi: 10.1146/annurev-genet-102108-134255. Annu Rev Genet. 2009. PMID: 19653859 Review.
Cited by
-
Metagenomic Analysis of Virus Diversity and Relative Abundance in a Eutrophic Freshwater Harbour.Viruses. 2019 Aug 28;11(9):792. doi: 10.3390/v11090792. Viruses. 2019. PMID: 31466255 Free PMC article.
-
In Situ Molecular Ecological Analyses Illuminate Distinct Factors Regulating Formation and Demise of a Harmful Dinoflagellate Bloom.Microbiol Spectr. 2023 Jun 15;11(3):e0515722. doi: 10.1128/spectrum.05157-22. Epub 2023 Apr 19. Microbiol Spectr. 2023. PMID: 37074171 Free PMC article.
-
Past and present giant viruses diversity explored through permafrost metagenomics.Nat Commun. 2022 Oct 7;13(1):5853. doi: 10.1038/s41467-022-33633-x. Nat Commun. 2022. PMID: 36207343 Free PMC article.
-
Mitochondrial Genome Evolution in Pelagophyte Algae.Genome Biol Evol. 2021 Mar 1;13(3):evab018. doi: 10.1093/gbe/evab018. Genome Biol Evol. 2021. PMID: 33675661 Free PMC article.
-
Giant virus biology and diversity in the era of genome-resolved metagenomics.Nat Rev Microbiol. 2022 Dec;20(12):721-736. doi: 10.1038/s41579-022-00754-5. Epub 2022 Jul 28. Nat Rev Microbiol. 2022. PMID: 35902763 Review.
References
-
- Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol Evol 4:1324–1330. doi:10.1093/gbe/evs109. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

