Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism
- PMID: 28442575
- PMCID: PMC5465478
- DOI: 10.1074/jbc.M117.785287
Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism
Abstract
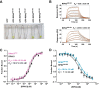
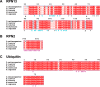
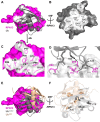
The 26S proteasome is a large cellular assembly that mediates the selective degradation of proteins in the nucleus and cytosol and is an established target for anticancer therapeutics. Protein substrates are typically targeted to the proteasome through modification with a polyubiquitin chain, which can be recognized by several proteasome-associated ubiquitin receptors. One of these receptors, RPN13/ADRM1, is recruited to the proteasome through direct interaction with the large scaffolding protein RPN2 within the 19S regulatory particle. To better understand the interactions between RPN13, RPN2, and ubiquitin, we used human proteins to map the RPN13-binding epitope to the C-terminal 14 residues of RPN2, which, like ubiquitin, binds the N-terminal pleckstrin-like receptor of ubiquitin (PRU) domain of RPN13. We also report the crystal structures of the RPN13 PRU domain in complex with peptides corresponding to the RPN2 C terminus and ubiquitin. Through mutational analysis, we validated the RPN2-binding interface revealed by our structures and quantified binding interactions with surface plasmon resonance and fluorescence polarization. In contrast to a previous report, we find that RPN13 binds ubiquitin with an affinity similar to that of other proteasome-associated ubiquitin receptors and that RPN2, ubiquitin, and the deubiquitylase UCH37 bind to RPN13 with independent energetics. These findings provide a detailed characterization of interactions that are important for proteasome function, indicate ubiquitin affinities that are consistent with the role of RPN13 as a proteasomal ubiquitin receptor, and have major implications for the development of novel anticancer therapeutics.
Keywords: RPN13; RPN2; crystallography; fluorescence polarization; proteasome; protein-protein interaction; surface plasmon resonance (SPR); ubiquitin.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.J Biol Chem. 2019 Jun 21;294(25):9659-9665. doi: 10.1074/jbc.AC119.008881. Epub 2019 May 7. J Biol Chem. 2019. PMID: 31064842 Free PMC article.
-
Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.Nature. 2008 May 22;453(7194):548-52. doi: 10.1038/nature06924. Nature. 2008. PMID: 18497827 Free PMC article.
-
Proteasome subunit Rpn13 is a novel ubiquitin receptor.Nature. 2008 May 22;453(7194):481-8. doi: 10.1038/nature06926. Nature. 2008. PMID: 18497817 Free PMC article.
-
Deubiquitination Reactions on the Proteasome for Proteasome Versatility.Int J Mol Sci. 2020 Jul 27;21(15):5312. doi: 10.3390/ijms21155312. Int J Mol Sci. 2020. PMID: 32726943 Free PMC article. Review.
-
Substrate receptors of proteasomes.Biol Rev Camb Philos Soc. 2018 Nov;93(4):1765-1777. doi: 10.1111/brv.12419. Epub 2018 May 6. Biol Rev Camb Philos Soc. 2018. PMID: 29732666 Review.
Cited by
-
Differential toxicity of ataxin-3 isoforms in Drosophila models of Spinocerebellar Ataxia Type 3.Neurobiol Dis. 2019 Dec;132:104535. doi: 10.1016/j.nbd.2019.104535. Epub 2019 Jul 13. Neurobiol Dis. 2019. PMID: 31310802 Free PMC article.
-
Mechanisms and regulation of substrate degradation by the 26S proteasome.Nat Rev Mol Cell Biol. 2025 Feb;26(2):104-122. doi: 10.1038/s41580-024-00778-0. Epub 2024 Oct 3. Nat Rev Mol Cell Biol. 2025. PMID: 39362999 Review.
-
Expression and Regulation of Deubiquitinase-Resistant, Unanchored Ubiquitin Chains in Drosophila.Sci Rep. 2018 May 31;8(1):8513. doi: 10.1038/s41598-018-26364-x. Sci Rep. 2018. PMID: 29855490 Free PMC article.
-
Discovery of a non-covalent ligand for Rpn-13, a therapeutic target for hematological cancers.Bioorg Med Chem Lett. 2023 Oct 15;95:129485. doi: 10.1016/j.bmcl.2023.129485. Epub 2023 Sep 14. Bioorg Med Chem Lett. 2023. PMID: 37714498 Free PMC article.
-
To Kill or to Be Killed: How Does the Battle between the UPS and Autophagy Maintain the Intracellular Homeostasis in Eukaryotes?Int J Mol Sci. 2023 Jan 22;24(3):2221. doi: 10.3390/ijms24032221. Int J Mol Sci. 2023. PMID: 36768543 Free PMC article. Review.
References
-
- Pickart C. M., and Cohen R. E. (2004) Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 - PubMed
-
- Fatimababy A. S., Lin Y. L., Usharani R., Radjacommare R., Wang H. T., Tsai H. L., Lee Y., and Fu H. (2010) Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 277, 796–816 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

