Evaluation of a rapid detection for Coxsackievirus B3 using one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP)
- PMID: 28435073
- PMCID: PMC7113869
- DOI: 10.1016/j.jviromet.2017.04.006
Evaluation of a rapid detection for Coxsackievirus B3 using one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP)
Abstract
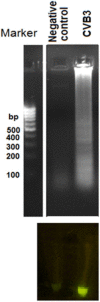
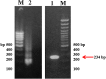
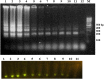
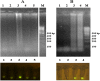
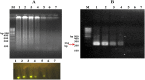
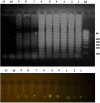
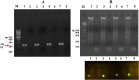
Coxsackievirus B3 (CVB3) is a member of the genus Enterovirus within the family Picornaviridae and is an important pathogen of viral myocarditis, which accounts for more than 50% viral myocarditis cases. VP1 is major capsid protein that this region has a low homology in both amino acid and nucleotide sequences among Enteroviruses. Therefore we have chosen this region for designed a set of RT-LAMP primers for CVB3 detection. For this the total RNA was extracted from 24-h post infected-HeLa cells with complete cytopathic effect (CPE), and applied to a one-step reverse transcription loop-mediated isothermal amplification reaction (RT-LAMP) using CVB3-specific primers. The optimization of RT-LAMP reaction was carried out with three variables factors including MgSO4 concentration, temperature and time of incubation. Amplification was analyzed by using 2% agarose gel electrophoresis and ethidium bromide and SYBR Green staining. Our results were shown the ladder-like pattern of the VP1 gene amplification. The LAMP reaction mix was optimized and the best result observed at 4mM MgSO4 and 60°C for 90min incubation. RT-LAMP had high sensitivity and specificity for detection of CVB3 infection. This method can be used as a rapid and easy diagnostic test for detection of CVB3 in clinical laboratories.
Keywords: Coxsackievirus B3; Detection; Optimization; RT-LAMP.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Development of a new method for diagnosis of Group B Coxsackie genome by reverse transcription loop-mediated isothermal amplification.Indian J Med Microbiol. 2011 Apr-Jun;29(2):110-7. doi: 10.4103/0255-0857.81780. Indian J Med Microbiol. 2011. PMID: 21654103
-
Detection of human Enterovirus 71 reverse transcription loop-mediated isothermal amplification (RT-LAMP).Lett Appl Microbiol. 2012 Mar;54(3):233-9. doi: 10.1111/j.1472-765X.2011.03198.x. Epub 2012 Jan 18. Lett Appl Microbiol. 2012. PMID: 22256968
-
Rapid and visual detection of human enterovirus coxsackievirus A16 by reverse transcription loop-mediated isothermal amplification combined with lateral flow device.Lett Appl Microbiol. 2015 Dec;61(6):531-7. doi: 10.1111/lam.12499. Epub 2015 Oct 30. Lett Appl Microbiol. 2015. PMID: 26399963
-
Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid detection of enterovirus 71.BMC Infect Dis. 2011 Jul 18;11:197. doi: 10.1186/1471-2334-11-197. BMC Infect Dis. 2011. PMID: 21767352 Free PMC article.
-
Characterisation of genomic RNA of Coxsackievirus B3 in murine myocarditis: reliability of direct sequencing of reverse transcription-nested polymerase chain reaction products.J Virol Methods. 1997 Dec;69(1-2):7-17. doi: 10.1016/s0166-0934(97)00122-5. J Virol Methods. 1997. PMID: 9504746
Cited by
-
One-step and Rapid Identification of SARS-CoV-2 using Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP).Avicenna J Med Biotechnol. 2024 Jan-Mar;16(1):3-8. doi: 10.18502/ajmb.v16i1.14165. Avicenna J Med Biotechnol. 2024. PMID: 38605744 Free PMC article.
-
Recent Developments in Isothermal Amplification Methods for the Detection of Foodborne Viruses.Front Microbiol. 2022 Mar 3;13:841875. doi: 10.3389/fmicb.2022.841875. eCollection 2022. Front Microbiol. 2022. PMID: 35308332 Free PMC article. Review.
-
Rapid molecular diagnosis of Parechovirus infection using the reverse transcription loop-mediated isothermal amplification technique.PLoS One. 2021 Nov 29;16(11):e0260348. doi: 10.1371/journal.pone.0260348. eCollection 2021. PLoS One. 2021. PMID: 34843518 Free PMC article.
-
Immunodiagnostic of Vibrio cholerae O1 using localized surface plasmon resonance (LSPR) biosensor.Int Microbiol. 2021 Jan;24(1):115-122. doi: 10.1007/s10123-020-00148-8. Epub 2020 Nov 4. Int Microbiol. 2021. PMID: 33150553
-
Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms.J Appl Microbiol. 2018 Mar;124(3):626-643. doi: 10.1111/jam.13647. Epub 2018 Feb 12. J Appl Microbiol. 2018. PMID: 29165905 Free PMC article. Review.
References
-
- Arita M., Ling H., Yan D., Nishimura Y., Yoshida H., Wakita T., Shimizu H. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) system for a highly sensitive detection of enterovirus in the stool samples of acute flaccid paralysis cases. BMC Infect. Dis. 2009;9(1) (p.1) - PMC - PubMed
-
- Bedard K.M., Semler B.L. Regulation of Picornavirus gene expression. Microbes Infect. 2004;6(7):702–713. - PubMed
-
- Caipang C.M., Haraguchi I., Ohira T., Hirono I., Aoki T. Rapid detection of a fish iridovirus using loop-mediated isothermal amplification (LAMP) J. Virol. Methods. 2004;121:155–161. - PubMed
-
- Chang G.H., Lin L., Luo Y.J., Cai L.J., Wu X.Y., Xu H.M., Zhu Q.Y. Sequence analysis of six enterovirus 71 strains with different virulences in humans. Virus Res. 2010;151(1):66–73. - PubMed
-
- Chen Q., Hu Z., Zhang Q., Yu M. Development and evaluation of a real-time method of simultaneous amplification and testing of enterovirus 71 incorporating a RNA internal control system. J. Virol. Methods. 2014;196:139–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

