Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration
- PMID: 28420437
- PMCID: PMC5395972
- DOI: 10.1186/s40478-017-0432-x
Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration
Abstract
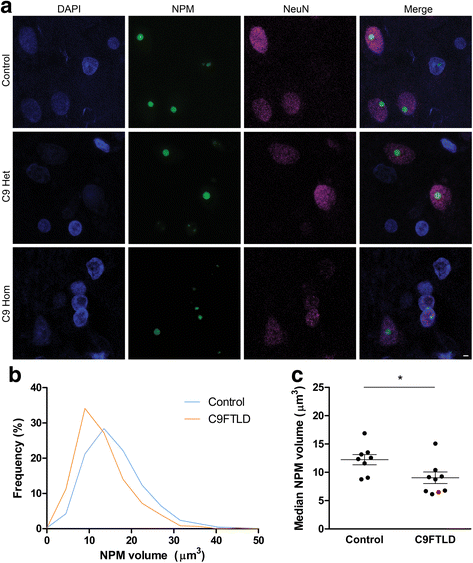
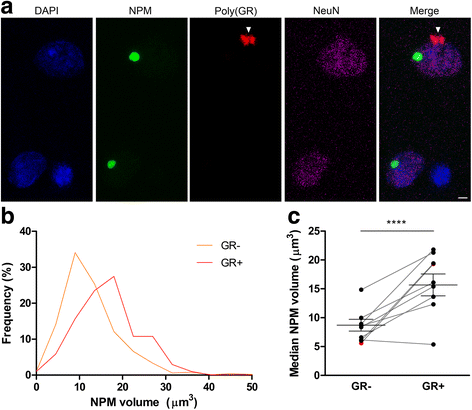
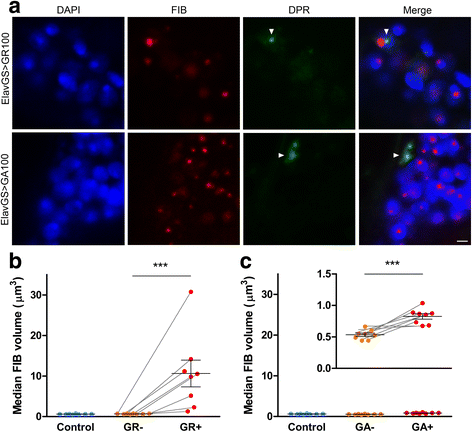
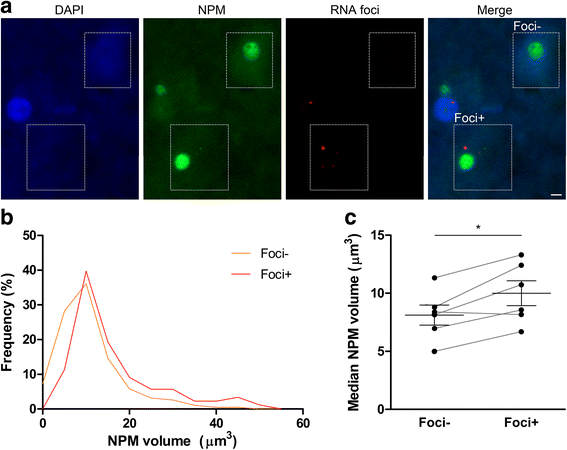
An intronic GGGGCC expansion in C9orf72 is the most common known cause of both frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). The repeat expansion leads to the generation of sense and antisense repeat RNA aggregates and dipeptide repeat (DPR) proteins, generated by repeat-associated non-ATG translation. The arginine-rich DPR proteins poly(glycine-arginine or GR) and poly(proline-arginine or PR) are potently neurotoxic and can localise to the nucleolus when expressed in cells, resulting in enlarged nucleoli with disrupted functionality. Furthermore, GGGGCC repeat RNA can bind nucleolar proteins in vitro. However, the relevance of nucleolar stress is unclear, as the arginine-rich DPR proteins do not localise to the nucleolus in C9orf72-associated FTLD/ALS (C9FTLD/ALS) patient brain. We measured nucleolar size in C9FTLD frontal cortex neurons using a three-dimensional, volumetric approach. Intriguingly, we found that C9FTLD brain exhibited bidirectional nucleolar stress. C9FTLD neuronal nucleoli were significantly smaller than control neuronal nucleoli. However, within C9FTLD brains, neurons containing poly(GR) inclusions had significantly larger nucleolar volumes than neurons without poly(GR) inclusions. In addition, expression of poly(GR) in adult Drosophila neurons led to significantly enlarged nucleoli. A small but significant increase in nucleolar volume was also observed in C9FTLD frontal cortex neurons containing GGGGCC repeat-containing RNA foci. These data show that nucleolar abnormalities are a consistent feature of C9FTLD brain, but that diverse pathomechanisms are at play, involving both DPR protein and repeat RNA toxicity.
Keywords: C9orf72; Dipeptide repeat proteins; FTLD; Nucleolar stress; Poly(GR); RNA foci.
Figures
Similar articles
-
Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing.Acta Neuropathol. 2015 Oct;130(4):537-55. doi: 10.1007/s00401-015-1450-z. Epub 2015 Jun 18. Acta Neuropathol. 2015. PMID: 26085200 Free PMC article.
-
C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci.Acta Neuropathol. 2013 Dec;126(6):845-57. doi: 10.1007/s00401-013-1200-z. Epub 2013 Oct 30. Acta Neuropathol. 2013. PMID: 24170096 Free PMC article.
-
Poly-GR dipeptide repeat polymers correlate with neurodegeneration and Clinicopathological subtypes in C9ORF72-related brain disease.Acta Neuropathol Commun. 2018 Jul 20;6(1):63. doi: 10.1186/s40478-018-0564-7. Acta Neuropathol Commun. 2018. PMID: 30029693 Free PMC article.
-
Mechanisms of toxicity in C9FTLD/ALS.Acta Neuropathol. 2014 Mar;127(3):359-76. doi: 10.1007/s00401-013-1237-z. Epub 2014 Jan 7. Acta Neuropathol. 2014. PMID: 24394885 Free PMC article. Review.
-
The neuropathology associated with repeat expansions in the C9ORF72 gene.Acta Neuropathol. 2014 Mar;127(3):347-57. doi: 10.1007/s00401-013-1232-4. Epub 2013 Dec 20. Acta Neuropathol. 2014. PMID: 24356984 Review.
Cited by
-
Altered Phase Separation and Cellular Impact in C9orf72-Linked ALS/FTD.Front Cell Neurosci. 2021 Apr 21;15:664151. doi: 10.3389/fncel.2021.664151. eCollection 2021. Front Cell Neurosci. 2021. PMID: 33967699 Free PMC article.
-
Poly-glycine-alanine exacerbates C9orf72 repeat expansion-mediated DNA damage via sequestration of phosphorylated ATM and loss of nuclear hnRNPA3.Acta Neuropathol. 2020 Jan;139(1):99-118. doi: 10.1007/s00401-019-02082-0. Epub 2019 Oct 23. Acta Neuropathol. 2020. PMID: 31642962 Free PMC article.
-
CRISPR-Cas9 Screens Identify the RNA Helicase DDX3X as a Repressor of C9ORF72 (GGGGCC)n Repeat-Associated Non-AUG Translation.Neuron. 2019 Dec 4;104(5):885-898.e8. doi: 10.1016/j.neuron.2019.09.003. Epub 2019 Oct 3. Neuron. 2019. PMID: 31587919 Free PMC article.
-
RNA-mediated toxicity in C9orf72 ALS and FTD.Neurobiol Dis. 2020 Nov;145:105055. doi: 10.1016/j.nbd.2020.105055. Epub 2020 Aug 21. Neurobiol Dis. 2020. PMID: 32829028 Free PMC article. Review.
-
New pathologic mechanisms in nucleotide repeat expansion disorders.Neurobiol Dis. 2019 Oct;130:104515. doi: 10.1016/j.nbd.2019.104515. Epub 2019 Jun 21. Neurobiol Dis. 2019. PMID: 31229686 Free PMC article. Review.
References
-
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, III, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. - DOI - PMC - PubMed
-
- Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, Campbell T, Uphill J, Borg A, Fratta P, Orrell RW, Malaspina A, Rowe J, Brown J, Hodges J, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. 2013;92:345–353. doi: 10.1016/j.ajhg.2013.01.011. - DOI - PMC - PubMed
-
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, Janke AM, Baatsen P, Vercruysse T, Kolaitis RM, Daelemans D, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055. doi: 10.1016/j.molcel.2017.02.013. - DOI - PMC - PubMed
-
- Cooper-Knock J, Walsh MJ, Higginbottom A, Robin HJ, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, Hautbergue GM, Shaw PJ. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–2051. doi: 10.1093/brain/awu120. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

