Structural homologies between phenformin, lipitor and gleevec aim the same metabolic oncotarget in leukemia and melanoma
- PMID: 28418852
- PMCID: PMC5564842
- DOI: 10.18632/oncotarget.16238
Structural homologies between phenformin, lipitor and gleevec aim the same metabolic oncotarget in leukemia and melanoma
Abstract
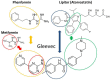
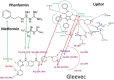
Phenformin's recently demonstrated efficacy in melanoma and Gleevec's demonstrated anti-proliferative action in chronic myeloid leukemia may lie within these drugs' significant pharmacokinetics, pharmacodynamics and structural homologies, which are reviewed herein. Gleevec's success in turning a fatal leukemia into a manageable chronic disease has been trumpeted in medical, economic, political and social circles because it is considered the first successful targeted therapy. Investments have been immense in omics analyses and while in some cases they greatly helped the management of patients, in others targeted therapies failed to achieve clinically stable recurrence-free disease course or to substantially extend survival. Nevertheless protein kinase controlling approaches have persisted despite early warnings that the targeted genomics narrative is overblown. Experimental and clinical observations with Phenformin suggest an alternative explanation for Gleevec's mode of action. Using 13C-guided precise flux measurements, a comparative multiple cell line study demonstrated the drug's downstream impact on submolecular fatty acid processing metabolic events that occurred independent of Gleevec's molecular target. Clinical observations that hyperlipidemia and diabetes are both reversed in mice and in patients taking Gleevec support the drugs' primary metabolic targets by biguanides and statins. This is evident by structural data demonstrating that Gleevec shows pyridine- and phenyl-guanidine homology with Phenformin and identical phenylcarbamoyl structural and ligand binding homology with Lipitor. The misunderstood mechanism of action of Gleevec is emblematic of the pervasive flawed reasoning that genomic analysis will lead to targeted, personalized diagnosis and therapy. The alternative perspective for Gleevec's mode of action may turn oncotargets towards metabolic channel reaction architectures in leukemia and melanoma, as well as in other cancers.
Keywords: deuterobolomics; imatinib; lipitor; metformin; phenformin.
Conflict of interest statement
Gábor Somlyai is employed by HYD, LLC. T. Que Collins is employed by and László G. Boros has academic consulting arrangements with 13Cignature 2Health Metabolic Clinic, Santa Monica, CA 90403, USA. Dr. Boros is an academic advisor of SiDMAP, LLC under FDA/OO/OFBA/OAGS/DAP branch work order HHSF223201610399A. None of the above entities are traded publically or share views necessarily with that of the authors.
Figures
Similar articles
-
Explaining why Gleevec is a specific and potent inhibitor of Abl kinase.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1664-9. doi: 10.1073/pnas.1214330110. Epub 2013 Jan 14. Proc Natl Acad Sci U S A. 2013. PMID: 23319661 Free PMC article.
-
Molecular dynamics simulations provide insights into the origin of gleevec's selectivity toward human tyrosine kinases.J Biomol Struct Dyn. 2019 Jul;37(10):2733-2744. doi: 10.1080/07391102.2018.1496139. Epub 2018 Nov 1. J Biomol Struct Dyn. 2019. PMID: 30052122
-
Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells.Nat Med. 2017 Oct;23(10):1234-1240. doi: 10.1038/nm.4399. Epub 2017 Sep 18. Nat Med. 2017. PMID: 28920959 Free PMC article.
-
Imatinib mesylate in chronic myeloid leukemia: frontline treatment and long-term outcomes.Expert Rev Anticancer Ther. 2016;16(3):273-8. doi: 10.1586/14737140.2016.1151356. Expert Rev Anticancer Ther. 2016. PMID: 26852913 Review.
-
Mechanisms of resistance to imatinib mesylate in Bcr-Abl-positive leukemias.Curr Opin Oncol. 2002 Nov;14(6):616-20. doi: 10.1097/00001622-200211000-00005. Curr Opin Oncol. 2002. PMID: 12409651 Review.
Cited by
-
Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia.Leukemia. 2022 Jan;36(1):1-12. doi: 10.1038/s41375-021-01416-w. Epub 2021 Sep 24. Leukemia. 2022. PMID: 34561557 Free PMC article. Review.
-
Clinicopathologic study of succinate-dehydrogenase-deficient gastrointestinal stromal tumors: A single-institutional experience in China.Medicine (Baltimore). 2017 Aug;96(32):e7668. doi: 10.1097/MD.0000000000007668. Medicine (Baltimore). 2017. PMID: 28796048 Free PMC article.
-
Evaluation of Poorly Soluble Drugs' Dissolution Rate by Laser Scattering in Different Water Isotopologues.Molecules. 2021 Jan 24;26(3):601. doi: 10.3390/molecules26030601. Molecules. 2021. PMID: 33498881 Free PMC article.
-
The Potential Effect of Metformin on Cancer: An Umbrella Review.Front Endocrinol (Lausanne). 2019 Sep 18;10:617. doi: 10.3389/fendo.2019.00617. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31620081 Free PMC article.
-
Comparative Proteomics of Dying and Surviving Cancer Cells Improves the Identification of Drug Targets and Sheds Light on Cell Life/Death Decisions.Mol Cell Proteomics. 2018 Jun;17(6):1144-1155. doi: 10.1074/mcp.RA118.000610. Epub 2018 Mar 23. Mol Cell Proteomics. 2018. PMID: 29572246 Free PMC article.
References
-
- Savage DG, Antman KH. Imatinib mesylate-a new oral targeted therapy. N Engl J Med. 2002;346:683–693. - PubMed
-
- Collins F, Galas D. A new five-year plan for the U.S. Human Genome Project. Science. 1993;262:43–46. - PubMed
-
- Morgan G, Aftimos P, Awada A. Current-day precision oncology: from cancer prevention, screening, drug development, and treatment - have we fallen short of the promise? Curr Opin Oncol. 2016;28:441–446. - PubMed
-
- Boros LG, Lee WN, Cascante M. Imatinib and chronic-phase leukemias. N Engl J Med. 2002;347:67–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

