The LIN28/let-7 Pathway in Cancer
- PMID: 28400788
- PMCID: PMC5368188
- DOI: 10.3389/fgene.2017.00031
The LIN28/let-7 Pathway in Cancer
Abstract
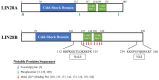
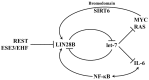
Among all tumor suppressor microRNAs, reduced let-7 expression occurs most frequently in cancer and typically correlates with poor prognosis. Activation of either LIN28A or LIN28B, two highly related RNA binding proteins (RBPs) and proto-oncogenes, is responsible for the global post-transcriptional downregulation of the let-7 microRNA family observed in many cancers. Specifically, LIN28A binds the terminal loop of precursor let-7 and recruits the Terminal Uridylyl Transferase (TUTase) ZCCHC11 that polyuridylates pre-let-7, thereby blocking microRNA biogenesis and tumor suppressor function. For LIN28B, the precise mechanism responsible for let-7 inhibition remains controversial. Functionally, the decrease in let-7 microRNAs leads to overexpression of their oncogenic targets such as MYC, RAS, HMGA2, BLIMP1, among others. Furthermore, mouse models demonstrate that ectopic LIN28 expression is sufficient to drive and/or accelerate tumorigenesis via a let-7 dependent mechanism. In this review, the LIN28/let-7 pathway is discussed, emphasizing its role in tumorigenesis, cancer stem cell biology, metabolomics, metastasis, and resistance to ionizing radiation and several chemotherapies. Also, emerging evidence will be presented suggesting that molecular targeting of this pathway may provide therapeutic benefit in cancer.
Keywords: Lin28; cancer stem cells; let-7; microRNAs; proto-oncogene proteins.
Figures



Similar articles
-
Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells.Nat Struct Mol Biol. 2009 Oct;16(10):1021-5. doi: 10.1038/nsmb.1676. Epub 2009 Aug 27. Nat Struct Mol Biol. 2009. PMID: 19713958 Free PMC article.
-
Identification of small molecule inhibitors of Zcchc11 TUTase activity.RNA Biol. 2015;12(8):792-800. doi: 10.1080/15476286.2015.1058478. RNA Biol. 2015. PMID: 26114892 Free PMC article.
-
Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7).RNA. 2012 Oct;18(10):1875-85. doi: 10.1261/rna.034538.112. Epub 2012 Aug 16. RNA. 2012. PMID: 22898984 Free PMC article.
-
Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer.Mol Cancer. 2015 Jun 30;14:125. doi: 10.1186/s12943-015-0402-5. Mol Cancer. 2015. PMID: 26123544 Free PMC article. Review.
-
3' RNA Uridylation in Epitranscriptomics, Gene Regulation, and Disease.Front Mol Biosci. 2018 Jul 13;5:61. doi: 10.3389/fmolb.2018.00061. eCollection 2018. Front Mol Biosci. 2018. PMID: 30057901 Free PMC article. Review.
Cited by
-
RNF144A suppresses ovarian cancer stem cell properties and tumor progression through regulation of LIN28B degradation via the ubiquitin-proteasome pathway.Cell Biol Toxicol. 2022 Oct;38(5):809-824. doi: 10.1007/s10565-021-09609-w. Epub 2021 May 11. Cell Biol Toxicol. 2022. PMID: 33978933
-
Emerging Clues of Regulatory Roles of Circular RNAs through Modulating Oxidative Stress: Focus on Neurological and Vascular Diseases.Oxid Med Cell Longev. 2021 Mar 1;2021:6659908. doi: 10.1155/2021/6659908. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 33747348 Free PMC article. Review.
-
The OTUD6B-LIN28B-MYC axis determines the proliferative state in multiple myeloma.EMBO J. 2022 Oct 17;41(20):e110871. doi: 10.15252/embj.2022110871. Epub 2022 Sep 5. EMBO J. 2022. PMID: 36059274 Free PMC article.
-
Malignancy-related mir-210, mir-373 and let-7 levels are affected in iron deficiency anemia.Afr Health Sci. 2023 Sep;23(3):245-253. doi: 10.4314/ahs.v23i3.30. Afr Health Sci. 2023. PMID: 38357103 Free PMC article.
-
RNA-Binding Proteins in Bladder Cancer.Cancers (Basel). 2023 Feb 10;15(4):1150. doi: 10.3390/cancers15041150. Cancers (Basel). 2023. PMID: 36831493 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

