Recent 5-year Findings and Technological Advances in the Proteomic Study of HIV-associated Disorders
- PMID: 28391008
- PMCID: PMC5415375
- DOI: 10.1016/j.gpb.2016.11.002
Recent 5-year Findings and Technological Advances in the Proteomic Study of HIV-associated Disorders
Abstract
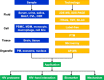
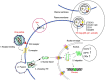
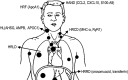
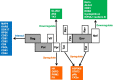
Human immunodeficiency virus-1 (HIV-1) mainly relies on host factors to complete its life cycle. Hence, it is very important to identify HIV-regulated host proteins. Proteomics is an excellent technique for this purpose because of its high throughput and sensitivity. In this review, we summarized current technological advances in proteomics, including general isobaric tags for relative and absolute quantitation (iTRAQ) and stable isotope labeling by amino acids in cell culture (SILAC), as well as subcellular proteomics and investigation of posttranslational modifications. Furthermore, we reviewed the applications of proteomics in the discovery of HIV-related diseases and HIV infection mechanisms. Proteins identified by proteomic studies might offer new avenues for the diagnosis and treatment of HIV infection and the related diseases.
Keywords: HIV-1; Interaction; Proteomics; SILAC; iTRAQ.
Copyright © 2017 The Authors. Production and hosting by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Topical capsaicin (high concentration) for chronic neuropathic pain in adults.Cochrane Database Syst Rev. 2017 Jan 13;1(1):CD007393. doi: 10.1002/14651858.CD007393.pub4. Cochrane Database Syst Rev. 2017. PMID: 28085183 Free PMC article. Review.
-
Australia in 2030: what is our path to health for all?Med J Aust. 2021 May;214 Suppl 8:S5-S40. doi: 10.5694/mja2.51020. Med J Aust. 2021. PMID: 33934362
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
The effects of radiofrequency electromagnetic field exposure on biomarkers of oxidative stress in vivo and in vitro: A systematic review of experimental studies.Environ Int. 2024 Dec;194:108940. doi: 10.1016/j.envint.2024.108940. Epub 2024 Aug 14. Environ Int. 2024. PMID: 39566441
Cited by
-
iTRAQ-based proteomics profiling of Schwann cells before and after peripheral nerve injury.Iran J Basic Med Sci. 2018 Aug;21(8):832-841. doi: 10.22038/IJBMS.2018.26944.6588. Iran J Basic Med Sci. 2018. PMID: 30186571 Free PMC article.
-
iTRAQ-based proteomic study on monocyte cell model discovered an association of LAMP2 downregulation with HIV-1 latency.Proteome Sci. 2024 May 15;22(1):6. doi: 10.1186/s12953-024-00230-3. Proteome Sci. 2024. PMID: 38750478 Free PMC article.
-
IFN-Inducible SerpinA5 Triggers Antiviral Immunity by Regulating STAT1 Phosphorylation and Nuclear Translocation.Int J Mol Sci. 2023 Mar 13;24(6):5458. doi: 10.3390/ijms24065458. Int J Mol Sci. 2023. PMID: 36982532 Free PMC article.
References
-
- Suguimoto S.P., Techasrivichien T., Musumari P.M., El-saaidi C., Lukhele B.W., Ono-Kihara M. Changing patterns of HIV epidemic in 30 years in East Asia. Curr HIV/AIDS Rep. 2014;11:134–145. - PubMed
-
- Bruun J.N., Skeie L., Maeland A., Dudman S.G., Sannes M., Ormaasen V. HIV/AIDS–from lethal syndrome to chronic disease. Tidsskr Nor Laegeforen. 2006;126:3121–3124. - PubMed
-
- Kumari G., Singh R.K. Anti-HIV drug development: structural features and limitations of present day drugs and future challenges in the successful HIV/AIDS treatment. Curr Pharm Des. 2013;19:1767–1783. - PubMed
-
- Este J.A., Cihlar T. Current status and challenges of antiretroviral research and therapy. Antiviral Res. 2010;85:25–33. - PubMed
-
- Clavel F., Hance A.J. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

