The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have
- PMID: 28371183
- PMCID: PMC6638112
- DOI: 10.1111/mpp.12553
The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have
Abstract
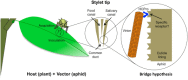
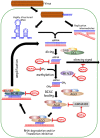
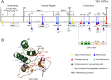
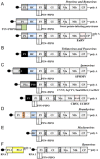
RNA viruses have very compact genomes and so provide a unique opportunity to study how evolution works to optimize the use of very limited genomic information. A widespread viral strategy to solve this issue concerning the coding space relies on the expression of proteins with multiple functions. Members of the family Potyviridae, the most abundant group of RNA viruses in plants, offer several attractive examples of viral factors which play roles in diverse infection-related pathways. The Helper Component Proteinase (HCPro) is an essential and well-characterized multitasking protein for which at least three independent functions have been described: (i) viral plant-to-plant transmission; (ii) polyprotein maturation; and (iii) RNA silencing suppression. Moreover, multitudes of host factors have been found to interact with HCPro. Intriguingly, most of these partners have not been ascribed to any of the HCPro roles during the infectious cycle, supporting the idea that this protein might play even more roles than those already established. In this comprehensive review, we attempt to summarize our current knowledge about HCPro and its already attributed and putative novel roles, and to discuss the similarities and differences regarding this factor in members of this important viral family.
Keywords: RNA silencing suppressor; multifunctional proteins; proteinase; transmission.
© 2017 BSPP AND JOHN WILEY & SONS LTD.
Figures
Similar articles
-
Molecular insights into the function of the viral RNA silencing suppressor HCPro.Plant J. 2016 Jan;85(1):30-45. doi: 10.1111/tpj.13088. Plant J. 2016. PMID: 26611351
-
A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles.J Virol. 2014 Sep 1;88(17):9808-18. doi: 10.1128/JVI.01010-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942578 Free PMC article.
-
Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif.J Virol. 2011 Jul;85(13):6784-94. doi: 10.1128/JVI.00485-11. Epub 2011 Apr 27. J Virol. 2011. PMID: 21525344 Free PMC article.
-
A Newly Identified Virus in the Family Potyviridae Encodes Two Leader Cysteine Proteases in Tandem That Evolved Contrasting RNA Silencing Suppression Functions.J Virol. 2020 Dec 9;95(1):e01414-20. doi: 10.1128/JVI.01414-20. Print 2020 Dec 9. J Virol. 2020. PMID: 33055249 Free PMC article.
-
The Biological Impact of the Hypervariable N-Terminal Region of Potyviral Genomes.Annu Rev Virol. 2019 Sep 29;6(1):255-274. doi: 10.1146/annurev-virology-092818-015843. Epub 2019 Jul 12. Annu Rev Virol. 2019. PMID: 31299166 Review.
Cited by
-
Maize Lethal Necrosis disease: review of molecular and genetic resistance mechanisms, socio-economic impacts, and mitigation strategies in sub-Saharan Africa.BMC Plant Biol. 2022 Nov 23;22(1):542. doi: 10.1186/s12870-022-03932-y. BMC Plant Biol. 2022. PMID: 36418954 Free PMC article. Review.
-
Adaptation of a Potyvirus Chimera Increases Its Virulence in a Compatible Host through Changes in HCPro.Plants (Basel). 2022 Aug 30;11(17):2262. doi: 10.3390/plants11172262. Plants (Basel). 2022. PMID: 36079643 Free PMC article.
-
Insight into aphid mediated Potato Virus Y transmission: A molecular to bioinformatics prospective.Front Microbiol. 2022 Nov 24;13:1001454. doi: 10.3389/fmicb.2022.1001454. eCollection 2022. Front Microbiol. 2022. PMID: 36504828 Free PMC article. Review.
-
Proteome expansion in the Potyviridae evolutionary radiation.FEMS Microbiol Rev. 2022 Jul 1;46(4):fuac011. doi: 10.1093/femsre/fuac011. FEMS Microbiol Rev. 2022. PMID: 35195244 Free PMC article. Review.
-
The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection.Viruses. 2020 Jan 8;12(1):77. doi: 10.3390/v12010077. Viruses. 2020. PMID: 31936267 Free PMC article. Review.
References
-
- Alamillo, J.M. , Sáenz, P. and García, J.A. (2006) Salicylic acid‐mediated and RNA‐silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 48, 217–227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

