Protein arginine methylation: a prominent modification and its demethylation
- PMID: 28364192
- PMCID: PMC11107486
- DOI: 10.1007/s00018-017-2515-z
Protein arginine methylation: a prominent modification and its demethylation
Abstract
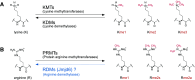
Arginine methylation of histones is one mechanism of epigenetic regulation in eukaryotic cells. Methylarginines can also be found in non-histone proteins involved in various different processes in a cell. An enzyme family of nine protein arginine methyltransferases catalyses the addition of methyl groups on arginines of histone and non-histone proteins, resulting in either mono- or dimethylated-arginine residues. The reversibility of histone modifications is an essential feature of epigenetic regulation to respond to changes in environmental factors, signalling events, or metabolic alterations. Prominent histone modifications like lysine acetylation and lysine methylation are reversible. Enzyme family pairs have been identified, with each pair of lysine acetyltransferases/deacetylases and lysine methyltransferases/demethylases operating complementarily to generate or erase lysine modifications. Several analyses also indicate a reversible nature of arginine methylation, but the enzymes facilitating direct removal of methyl moieties from arginine residues in proteins have been discussed controversially. Differing reports have been seen for initially characterized putative candidates, like peptidyl arginine deiminase 4 or Jumonji-domain containing protein 6. Here, we review the most recent cellular, biochemical, and mass spectrometry work on arginine methylation and its reversible nature with a special focus on putative arginine demethylases, including the enzyme superfamily of Fe(II) and 2-oxoglutarate-dependent oxygenases.
Keywords: Histone modifications; KDM; KDM2A; KDM3A; KDM4E; KDM5C; KDM6B; KDM7B; KMT; Liquid chromatography–tandem mass spectrometry; PHF8; Post-translational modifications.
Figures

Similar articles
-
The Kdm/Kmt gene families in the self-fertilizing mangrove rivulus fish, Kryptolebias marmoratus, suggest involvement of histone methylation machinery in development and reproduction.Gene. 2019 Mar 1;687:173-187. doi: 10.1016/j.gene.2018.11.046. Epub 2018 Nov 17. Gene. 2019. PMID: 30458291
-
Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review).Mol Med Rep. 2019 May;19(5):3963-3971. doi: 10.3892/mmr.2019.10111. Epub 2019 Apr 1. Mol Med Rep. 2019. PMID: 30942418 Free PMC article. Review.
-
HDAC inhibitors induce global changes in histone lysine and arginine methylation and alter expression of lysine demethylases.J Proteomics. 2016 Feb 5;133:125-133. doi: 10.1016/j.jprot.2015.12.018. Epub 2015 Dec 22. J Proteomics. 2016. PMID: 26721445
-
Intricate Effects of α-Amino and Lysine Modifications on Arginine Methylation of the N-Terminal Tail of Histone H4.Biochemistry. 2017 Jul 18;56(28):3539-3548. doi: 10.1021/acs.biochem.7b00450. Epub 2017 Jul 7. Biochemistry. 2017. PMID: 28644004 Free PMC article.
-
[Advances in histone methyltransferases and histone demethylases].Ai Zheng. 2008 Oct;27(10):1018-25. Ai Zheng. 2008. PMID: 18851779 Review. Chinese.
Cited by
-
R-Methylation in Plants: A Key Regulator of Plant Development and Response to the Environment.Int J Mol Sci. 2024 Sep 14;25(18):9937. doi: 10.3390/ijms25189937. Int J Mol Sci. 2024. PMID: 39337424 Free PMC article. Review.
-
Protein arginine methyltransferase 1 mediates renal fibroblast activation and fibrogenesis through activation of Smad3 signaling.Am J Physiol Renal Physiol. 2020 Feb 1;318(2):F375-F387. doi: 10.1152/ajprenal.00487.2019. Epub 2019 Dec 9. Am J Physiol Renal Physiol. 2020. PMID: 31813251 Free PMC article.
-
PRMT5 Facilitates Infectious Bursal Disease Virus Replication through Arginine Methylation of VP1.J Virol. 2023 Mar 30;97(3):e0163722. doi: 10.1128/jvi.01637-22. Epub 2023 Feb 14. J Virol. 2023. PMID: 36786602 Free PMC article.
-
The RGG motif proteins: Interactions, functions, and regulations.Wiley Interdiscip Rev RNA. 2023 Jan;14(1):e1748. doi: 10.1002/wrna.1748. Epub 2022 Jun 3. Wiley Interdiscip Rev RNA. 2023. PMID: 35661420 Free PMC article. Review.
-
[Transcription of protein arginine N-methyltransferase genes in mouse dorsal root ganglia following peripheral nerve injury].Nan Fang Yi Ke Da Xue Xue Bao. 2017 Dec 20;37(12):1620-1625. doi: 10.3969/j.issn.1673-4254.2017.12.10. Nan Fang Yi Ke Da Xue Xue Bao. 2017. PMID: 29292255 Free PMC article. Chinese.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

