Phosphatidylserine Lateral Organization Influences the Interaction of Influenza Virus Matrix Protein 1 with Lipid Membranes
- PMID: 28356535
- PMCID: PMC5446629
- DOI: 10.1128/JVI.00267-17
Phosphatidylserine Lateral Organization Influences the Interaction of Influenza Virus Matrix Protein 1 with Lipid Membranes
Abstract
Influenza A virus matrix protein 1 (M1) is an essential component involved in the structural stability of the virus and in the budding of new virions from infected cells. A deeper understanding of the molecular basis of virion formation and the budding process is required in order to devise new therapeutic approaches. We performed a detailed investigation of the interaction between M1 and phosphatidylserine (PS) (i.e., its main binding target at the plasma membrane [PM]), as well as the distribution of PS itself, both in model membranes and in living cells. To this end, we used a combination of techniques, including Förster resonance energy transfer (FRET), confocal microscopy imaging, raster image correlation spectroscopy, and number and brightness (N&B) analysis. Our results show that PS can cluster in segregated regions in the plane of the lipid bilayer, both in model bilayers constituted of PS and phosphatidylcholine and in living cells. The viral protein M1 interacts specifically with PS-enriched domains, and such interaction in turn affects its oligomerization process. Furthermore, M1 can stabilize PS domains, as observed in model membranes. For living cells, the presence of PS clusters is suggested by N&B experiments monitoring the clustering of the PS sensor lactadherin. Also, colocalization between M1 and a fluorescent PS probe suggest that, in infected cells, the matrix protein can specifically bind to the regions of PM in which PS is clustered. Taken together, our observations provide novel evidence regarding the role of PS-rich domains in tuning M1-lipid and M1-M1 interactions at the PM of infected cells.IMPORTANCE Influenza virus particles assemble at the plasma membranes (PM) of infected cells. This process is orchestrated by the matrix protein M1, which interacts with membrane lipids while binding to the other proteins and genetic material of the virus. Despite its importance, the initial step in virus assembly (i.e., M1-lipid interaction) is still not well understood. In this work, we show that phosphatidylserine can form lipid domains in physical models of the inner leaflet of the PM. Furthermore, the spatial organization of PS in the plane of the bilayer modulates M1-M1 interactions. Finally, we show that PS domains appear to be present in the PM of living cells and that M1 seems to display a high affinity for them.
Keywords: assembly; confocal microscopy; fluorescence image analysis; influenza; lipid rafts; matrix protein; model membranes; phosphatidylserine; plasma membrane.
Copyright © 2017 American Society for Microbiology.
Figures
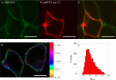
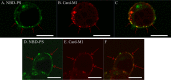
Similar articles
-
Lateral Organization of Influenza Virus Proteins in the Budozone Region of the Plasma Membrane.J Virol. 2017 Apr 13;91(9):e02104-16. doi: 10.1128/JVI.02104-16. Print 2017 May 1. J Virol. 2017. PMID: 28202765 Free PMC article.
-
Influenza A matrix protein M1 induces lipid membrane deformation via protein multimerization.Biosci Rep. 2019 Aug 5;39(8):BSR20191024. doi: 10.1042/BSR20191024. Print 2019 Aug 30. Biosci Rep. 2019. PMID: 31324731 Free PMC article.
-
Influenza A matrix protein M1 multimerizes upon binding to lipid membranes.Biophys J. 2014 Aug 19;107(4):912-23. doi: 10.1016/j.bpj.2014.06.042. Biophys J. 2014. PMID: 25140426 Free PMC article.
-
Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses.Virus Res. 2001 Sep;77(1):61-9. doi: 10.1016/s0168-1702(01)00266-0. Virus Res. 2001. PMID: 11451488 Review.
-
Assembly and budding of influenza virus.Virus Res. 2004 Dec;106(2):147-65. doi: 10.1016/j.virusres.2004.08.012. Virus Res. 2004. PMID: 15567494 Free PMC article. Review.
Cited by
-
Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies.PLoS One. 2020 Oct 15;15(10):e0240653. doi: 10.1371/journal.pone.0240653. eCollection 2020. PLoS One. 2020. PMID: 33057452 Free PMC article.
-
Phosphatidylinositol 4,5-Bisphosphate Mediates the Co-Distribution of Influenza A Hemagglutinin and Matrix Protein M1 at the Plasma Membrane.Viruses. 2022 Nov 12;14(11):2509. doi: 10.3390/v14112509. Viruses. 2022. PMID: 36423118 Free PMC article.
-
Physico-Chemical Mechanisms of the Functioning of Membrane-Active Proteins of Enveloped Viruses.Biochem (Mosc) Suppl Ser A Membr Cell Biol. 2022;16(4):247-260. doi: 10.1134/S1990747822050038. Epub 2022 Dec 9. Biochem (Mosc) Suppl Ser A Membr Cell Biol. 2022. PMID: 36532264 Free PMC article.
-
Influenza A Virus M1 Protein Non-Specifically Deforms Charged Lipid Membranes and Specifically Interacts with the Raft Boundary.Membranes (Basel). 2023 Jan 7;13(1):76. doi: 10.3390/membranes13010076. Membranes (Basel). 2023. PMID: 36676883 Free PMC article.
-
Graph-Based Analyses of Dynamic Water-Mediated Hydrogen-Bond Networks in Phosphatidylserine: Cholesterol Membranes.Biomolecules. 2023 Aug 11;13(8):1238. doi: 10.3390/biom13081238. Biomolecules. 2023. PMID: 37627303 Free PMC article.
References
-
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi:10.1038/nm1477. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

