Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size
- PMID: 28351851
- PMCID: PMC5412557
- DOI: 10.1083/jcb.201607022
Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size
Abstract
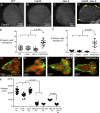
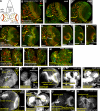
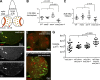
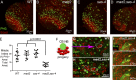
Accurate mitotic spindle assembly is critical for mitotic fidelity and organismal development. Multiple processes coordinate spindle assembly and chromosome segregation. Two key components are centrosomes and the spindle assembly checkpoint (SAC), and mutations affecting either can cause human microcephaly. In vivo studies in Drosophila melanogaster found that loss of either component alone is well tolerated in the developing brain, in contrast to epithelial tissues of the imaginal discs. In this study, we reveal that one reason for that tolerance is the compensatory relationship between centrosomes and the SAC. In the absence of both centrosomes and the SAC, brain cells, including neural stem cells, experience massive errors in mitosis, leading to increased cell death, which reduces the neural progenitor pool and severely disrupts brain development. However, our data also demonstrate that neural cells are much more tolerant of aneuploidy than epithelial cells. Our data provide novel insights into the mechanisms by which different tissues manage genome stability and parallels with human microcephaly.
© 2017 Poulton et al.
Figures
Similar articles
-
Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size.Nat Commun. 2014 May 30;5:3885. doi: 10.1038/ncomms4885. Nat Commun. 2014. PMID: 24875059 Free PMC article.
-
Centrosomes and the art of mitotic spindle maintenance.Int Rev Cell Mol Biol. 2014;313:179-217. doi: 10.1016/B978-0-12-800177-6.00006-2. Int Rev Cell Mol Biol. 2014. PMID: 25376493 Review.
-
Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity.Cell. 2003 Apr 4;113(1):87-99. doi: 10.1016/s0092-8674(03)00202-2. Cell. 2003. PMID: 12679037
-
Centrosomes: The good and the bad for brain development.Biol Cell. 2020 Jun;112(6):153-172. doi: 10.1111/boc.201900090. Epub 2020 Mar 26. Biol Cell. 2020. PMID: 32170757 Review.
-
Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation.Dev Cell. 2014 Sep 29;30(6):731-45. doi: 10.1016/j.devcel.2014.08.007. Epub 2014 Sep 18. Dev Cell. 2014. PMID: 25241934 Free PMC article.
Cited by
-
Genomic instability and cancer: lessons from Drosophila.Open Biol. 2020 Jun;10(6):200060. doi: 10.1098/rsob.200060. Epub 2020 Jun 3. Open Biol. 2020. PMID: 32485126 Free PMC article. Review.
-
Altered cleavage plane orientation with increased genomic aneuploidy produced by receptor-mediated lysophosphatidic acid (LPA) signaling in mouse cerebral cortical neural progenitor cells.Mol Brain. 2020 Dec 14;13(1):169. doi: 10.1186/s13041-020-00709-y. Mol Brain. 2020. PMID: 33317583 Free PMC article.
-
Traip controls mushroom body size by suppressing mitotic defects.Development. 2022 Apr 1;149(7):dev199987. doi: 10.1242/dev.199987. Epub 2022 Mar 31. Development. 2022. PMID: 35297981 Free PMC article.
-
Chromosome integrity checkpoints in stem and progenitor cells: transitions upon differentiation, pathogenesis, and aging.Cell Mol Life Sci. 2018 Oct;75(20):3771-3779. doi: 10.1007/s00018-018-2891-z. Epub 2018 Jul 31. Cell Mol Life Sci. 2018. PMID: 30066086 Free PMC article. Review.
-
The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases.Cells. 2019 Dec 24;9(1):49. doi: 10.3390/cells9010049. Cells. 2019. PMID: 31878213 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

