Pharmacogenomic identification of small molecules for lineage specific manipulation of subventricular zone germinal activity
- PMID: 28350803
- PMCID: PMC5370089
- DOI: 10.1371/journal.pbio.2000698
Pharmacogenomic identification of small molecules for lineage specific manipulation of subventricular zone germinal activity
Abstract
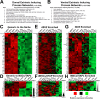
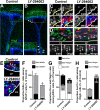
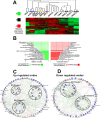
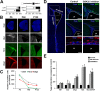
Strategies for promoting neural regeneration are hindered by the difficulty of manipulating desired neural fates in the brain without complex genetic methods. The subventricular zone (SVZ) is the largest germinal zone of the forebrain and is responsible for the lifelong generation of interneuron subtypes and oligodendrocytes. Here, we have performed a bioinformatics analysis of the transcriptome of dorsal and lateral SVZ in early postnatal mice, including neural stem cells (NSCs) and their immediate progenies, which generate distinct neural lineages. We identified multiple signaling pathways that trigger distinct downstream transcriptional networks to regulate the diversity of neural cells originating from the SVZ. Next, we used a novel in silico genomic analysis, searchable platform-independent expression database/connectivity map (SPIED/CMAP), to generate a catalogue of small molecules that can be used to manipulate SVZ microdomain-specific lineages. Finally, we demonstrate that compounds identified in this analysis promote the generation of specific cell lineages from NSCs in vivo, during postnatal life and adulthood, as well as in regenerative contexts. This study unravels new strategies for using small bioactive molecules to direct germinal activity in the SVZ, which has therapeutic potential in neurodegenerative diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Persistent Wnt/β-catenin signaling determines dorsalization of the postnatal subventricular zone and neural stem cell specification into oligodendrocytes and glutamatergic neurons.Stem Cells. 2014 May;32(5):1301-12. doi: 10.1002/stem.1639. Stem Cells. 2014. PMID: 24449255
-
HOPX Defines Heterogeneity of Postnatal Subventricular Zone Neural Stem Cells.Stem Cell Reports. 2018 Sep 11;11(3):770-783. doi: 10.1016/j.stemcr.2018.08.006. Epub 2018 Aug 30. Stem Cell Reports. 2018. PMID: 30174314 Free PMC article.
-
Transcriptional Hallmarks of Heterogeneous Neural Stem Cell Niches of the Subventricular Zone.Stem Cells. 2015 Jul;33(7):2232-42. doi: 10.1002/stem.2017. Epub 2015 Apr 23. Stem Cells. 2015. PMID: 25827345
-
The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis.Cold Spring Harb Perspect Biol. 2016 May 2;8(5):a018820. doi: 10.1101/cshperspect.a018820. Cold Spring Harb Perspect Biol. 2016. PMID: 27048191 Free PMC article. Review.
-
Role of the nuclear receptor Tailless in adult neural stem cells.Mech Dev. 2013 Jun-Aug;130(6-8):388-90. doi: 10.1016/j.mod.2013.02.001. Epub 2013 Feb 13. Mech Dev. 2013. PMID: 23415832 Review.
Cited by
-
Amburana cearensis: Pharmacological and Neuroprotective Effects of Its Compounds.Molecules. 2020 Jul 27;25(15):3394. doi: 10.3390/molecules25153394. Molecules. 2020. PMID: 32726999 Free PMC article. Review.
-
The Genomic Intersection of Oligodendrocyte Dynamics in Schizophrenia and Aging Unravels Novel Pathological Mechanisms and Therapeutic Potentials.Int J Mol Sci. 2024 Apr 18;25(8):4452. doi: 10.3390/ijms25084452. Int J Mol Sci. 2024. PMID: 38674040 Free PMC article. Review.
-
Gender-specific effects of transthyretin on neural stem cell fate in the subventricular zone of the adult mouse.Sci Rep. 2019 Dec 23;9(1):19689. doi: 10.1038/s41598-019-56156-w. Sci Rep. 2019. PMID: 31873158 Free PMC article.
-
Astrocytes are direct cellular targets of lithium treatment: novel roles for lysyl oxidase and peroxisome-proliferator activated receptor-γ as astroglial targets of lithium.Transl Psychiatry. 2019 Sep 2;9(1):211. doi: 10.1038/s41398-019-0542-2. Transl Psychiatry. 2019. PMID: 31477687 Free PMC article.
-
Drug repurposing for neuroregeneration in multiple sclerosis.Neural Regen Res. 2018 Aug;13(8):1366-1367. doi: 10.4103/1673-5374.235242. Neural Regen Res. 2018. PMID: 30106047 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

