Towards reproducible MRM based biomarker discovery using dried blood spots
- PMID: 28345601
- PMCID: PMC5366927
- DOI: 10.1038/srep45178
Towards reproducible MRM based biomarker discovery using dried blood spots
Abstract
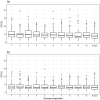
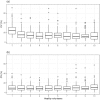
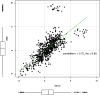
There is an increasing interest in the use of dried blood spot (DBS) sampling and multiple reaction monitoring in proteomics. Although several groups have explored the utility of DBS by focusing on protein detection, the reproducibility of the approach and whether it can be used for biomarker discovery in high throughput studies is yet to be determined. We assessed the reproducibility of multiplexed targeted protein measurements in DBS compared to serum. Eighty-two medium to high abundance proteins were monitored in a number of technical and biological replicates. Importantly, as part of the data analysis, several statistical quality control approaches were evaluated to detect inaccurate transitions. After implementing statistical quality control measures, the median CV on the original scale for all detected peptides in DBS was 13.2% and in Serum 8.8%. We also found a strong correlation (r = 0.72) between relative peptide abundance measured in DBS and serum. The combination of minimally invasive sample collection with a highly specific and sensitive mass spectrometry (MS) technique allows for targeted quantification of multiple proteins in a single MS run. This approach has the potential to fundamentally change clinical proteomics and personalized medicine by facilitating large-scale studies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Multiplexed quantitation of endogenous proteins in dried blood spots by multiple reaction monitoring-mass spectrometry.Mol Cell Proteomics. 2013 Mar;12(3):781-91. doi: 10.1074/mcp.M112.022442. Epub 2012 Dec 7. Mol Cell Proteomics. 2013. PMID: 23221968 Free PMC article.
-
Multiple Reaction Monitoring Enables Precise Quantification of 97 Proteins in Dried Blood Spots.Mol Cell Proteomics. 2015 Nov;14(11):3094-104. doi: 10.1074/mcp.O115.049957. Epub 2015 Sep 4. Mol Cell Proteomics. 2015. PMID: 26342038 Free PMC article.
-
Quantitation of therapeutic proteins following direct trypsin digestion of dried blood spot samples and detection by LC-MS-based bioanalytical methods in drug discovery.Bioanalysis. 2012 Jan;4(1):29-40. doi: 10.4155/bio.11.293. Bioanalysis. 2012. PMID: 22191592
-
Emerging liquid chromatography-mass spectrometry technologies improving dried blood spot analysis.Expert Rev Proteomics. 2014 Aug;11(4):425-30. doi: 10.1586/14789450.2014.904204. Epub 2014 Apr 3. Expert Rev Proteomics. 2014. PMID: 24697571 Review.
-
The use of mass spectrometry to analyze dried blood spots.Mass Spectrom Rev. 2016 May-Jun;35(3):361-438. doi: 10.1002/mas.21441. Epub 2014 Sep 22. Mass Spectrom Rev. 2016. PMID: 25252132 Review.
Cited by
-
Internal Standard Triggered-Parallel Reaction Monitoring Mass Spectrometry Enables Multiplexed Quantification of Candidate Biomarkers in Plasma.Anal Chem. 2022 Jul 12;94(27):9540-9547. doi: 10.1021/acs.analchem.1c04382. Epub 2022 Jun 29. Anal Chem. 2022. PMID: 35767427 Free PMC article.
-
Multimodel inference for biomarker development: an application to schizophrenia.Transl Psychiatry. 2019 Feb 11;9(1):83. doi: 10.1038/s41398-019-0419-4. Transl Psychiatry. 2019. PMID: 30745560 Free PMC article.
-
Use of a Novel Whole Blood Separation and Transport Device for Targeted and Untargeted Proteomics.Biomedicines. 2024 Oct 11;12(10):2318. doi: 10.3390/biomedicines12102318. Biomedicines. 2024. PMID: 39457630 Free PMC article.
-
A Combined Digital and Biomarker Diagnostic Aid for Mood Disorders (the Delta Trial): Protocol for an Observational Study.JMIR Res Protoc. 2020 Aug 10;9(8):e18453. doi: 10.2196/18453. JMIR Res Protoc. 2020. PMID: 32773373 Free PMC article.
-
A proposal for score assignment to characterize biological processes from mass spectral analysis of serum.Clin Mass Spectrom. 2020 Sep 9;18:13-26. doi: 10.1016/j.clinms.2020.09.001. eCollection 2020 Nov. Clin Mass Spectrom. 2020. PMID: 34820522 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

