Translation Initiation Factor eIF4E and eIFiso4E Are Both Required for Peanut stripe virus Infection in Peanut (Arachis hypogaea L.)
- PMID: 28344571
- PMCID: PMC5344889
- DOI: 10.3389/fmicb.2017.00338
Translation Initiation Factor eIF4E and eIFiso4E Are Both Required for Peanut stripe virus Infection in Peanut (Arachis hypogaea L.)
Abstract
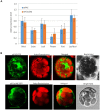
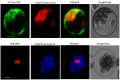
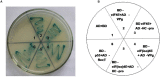
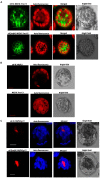
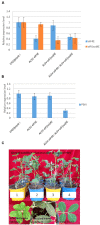
Peanut stripe virus (PStV) belongs to the genus Potyvirus and is the most important viral pathogen of cultivated peanut (Arachis hypogaea L.). The eukaryotic translation initiation factor, eIF4E, and its isoform, eIF(iso)4E, play key roles during virus infection in plants, particularly Potyvirus. In the present study, we cloned the eIF4E and eIF(iso)4E homologs in peanut and named these as PeaeIF4E and PeaeIF(iso)4E, respectively. Quantitative real-time PCR (qRT-PCR) analysis showed that these two genes were expressed during all growth periods and in all peanut organs, but were especially abundant in young leaves and roots. These also had similar expression levels. Yeast two-hybrid analysis showed that PStV multifunctional helper component proteinase (HC-Pro) and viral protein genome-linked (VPg) both interacted with PeaeIF4E and PeaeIF(iso)4E. Bimolecular fluorescence complementation assay showed that there was an interaction between HC-Pro and PeaeIF4E/PeaeIF(iso)4E in the cytoplasm and between VPg and PeaeIF4E/PeaeIF(iso)4E in the nucleus. Silencing either PeaeIF4E or PeaeIF(iso)4E using a virus-induced gene silencing system did not significantly affect PStV accumulation. However, silencing both PeaeIF4E and PeaeIF(iso)4E genes significantly weakened PStV accumulation. The findings of the present study suggest that PeaeIF4E and PeaeIF(iso)4E play important roles in the PStV infection cycle and may potentially contribute to PStV resistance.
Keywords: Peanut stripe virus; gene silencing; peanut; protein–protein interaction; translation initiation factor 4E.
Figures
Similar articles
-
Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif.J Virol. 2011 Jul;85(13):6784-94. doi: 10.1128/JVI.00485-11. Epub 2011 Apr 27. J Virol. 2011. PMID: 21525344 Free PMC article.
-
Variability in eukaryotic initiation factor iso4E in Brassica rapa influences interactions with the viral protein linked to the genome of Turnip mosaic virus.Sci Rep. 2018 Sep 11;8(1):13588. doi: 10.1038/s41598-018-31739-1. Sci Rep. 2018. PMID: 30206242 Free PMC article.
-
Selective Interaction of Sugarcane eIF4E with VPgs from Sugarcane Mosaic Pathogens.Viruses. 2021 Mar 22;13(3):518. doi: 10.3390/v13030518. Viruses. 2021. PMID: 33809985 Free PMC article.
-
A Novel Interaction Network Used by Potyviruses in Virus-Host Interactions at the Protein Level.Viruses. 2019 Dec 14;11(12):1158. doi: 10.3390/v11121158. Viruses. 2019. PMID: 31847316 Free PMC article.
-
Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement.Mol Plant Pathol. 2012 Sep;13(7):795-803. doi: 10.1111/j.1364-3703.2012.00791.x. Epub 2012 Mar 2. Mol Plant Pathol. 2012. PMID: 22379950 Free PMC article. Review.
Cited by
-
Advances in omics research on peanut response to biotic stresses.Front Plant Sci. 2023 May 22;14:1101994. doi: 10.3389/fpls.2023.1101994. eCollection 2023. Front Plant Sci. 2023. PMID: 37284721 Free PMC article. Review.
-
Potyviral Helper-Component Protease: Multifaced Functions and Interactions with Host Proteins.Plants (Basel). 2024 Apr 29;13(9):1236. doi: 10.3390/plants13091236. Plants (Basel). 2024. PMID: 38732454 Free PMC article. Review.
-
Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance.Mol Plant Pathol. 2020 Mar;21(3):303-317. doi: 10.1111/mpp.12897. Epub 2019 Dec 20. Mol Plant Pathol. 2020. PMID: 31860775 Free PMC article.
-
AhABI4s Negatively Regulate Salt-Stress Response in Peanut.Front Plant Sci. 2021 Oct 14;12:741641. doi: 10.3389/fpls.2021.741641. eCollection 2021. Front Plant Sci. 2021. PMID: 34721468 Free PMC article.
-
Eukaryotic translation initiation factor 4E family member nCBP facilitates the accumulation of TGB-encoding viruses by recognizing the viral coat protein in potato and tobacco.Front Plant Sci. 2022 Aug 8;13:946873. doi: 10.3389/fpls.2022.946873. eCollection 2022. Front Plant Sci. 2022. PMID: 36003826 Free PMC article.
References
-
- Ala-Poikela M., Goytia E., Haikonen T., Rajamaki M. L., Valkonen J. P. (2011). Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85 6784–6794. 10.1128/JVI.00485-11 - DOI - PMC - PubMed
-
- Bian L. (2013). Structural and Subcellular Distribution of VPg Protein Encode by Wheat Yellow Masaic Virus. Master thesis, Zhejiang Normal University, Jinhua.
LinkOut - more resources
Full Text Sources
Other Literature Sources

