Concordance analysis of microarray studies identifies representative gene expression changes in Parkinson's disease: a comparison of 33 human and animal studies
- PMID: 28335819
- PMCID: PMC5364698
- DOI: 10.1186/s12883-017-0838-x
Concordance analysis of microarray studies identifies representative gene expression changes in Parkinson's disease: a comparison of 33 human and animal studies
Erratum in
-
Correction to: Concordance analysis of microarray studies identifies representative gene expression changes in Parkinson's disease: a comparison of 33 human and animal studies.BMC Neurol. 2019 Jan 30;19(1):16. doi: 10.1186/s12883-019-1240-7. BMC Neurol. 2019. PMID: 30700273 Free PMC article.
Abstract
Background: As the popularity of transcriptomic analysis has grown, the reported lack of concordance between different studies of the same condition has become a growing concern, raising questions as to the representativeness of different study types, such as non-human disease models or studies of surrogate tissues, to gene expression in the human condition.
Methods: In a comparison of 33 microarray studies of Parkinson's disease, correlation and clustering analyses were used to determine the factors influencing concordance between studies, including agreement between different tissue types, different microarray platforms, and between neurotoxic and genetic disease models and human Parkinson's disease.
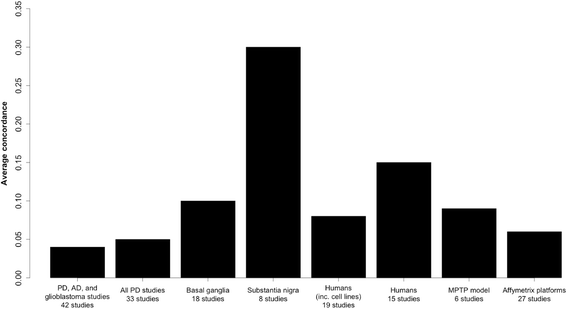
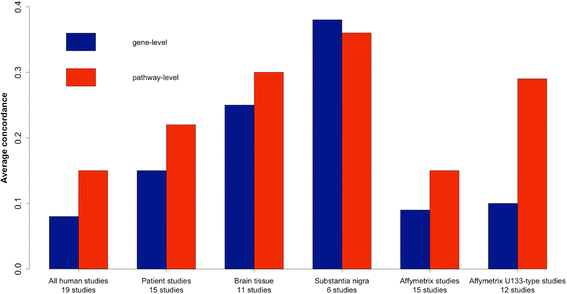
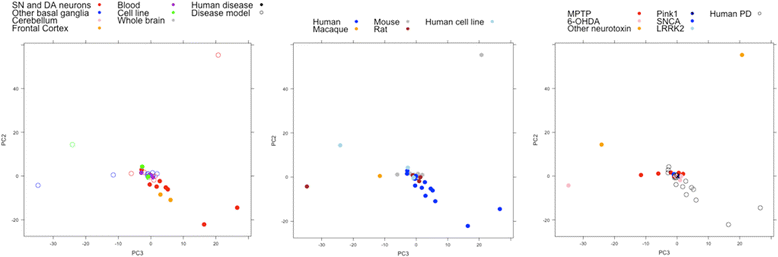
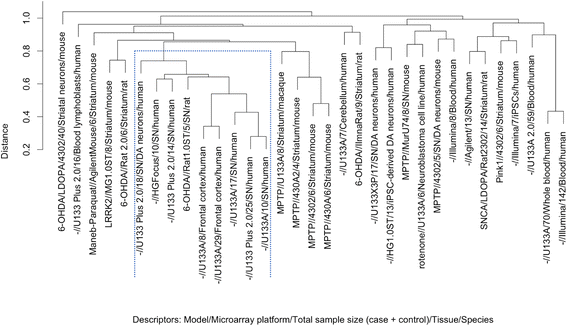
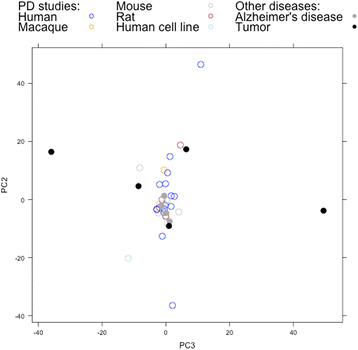
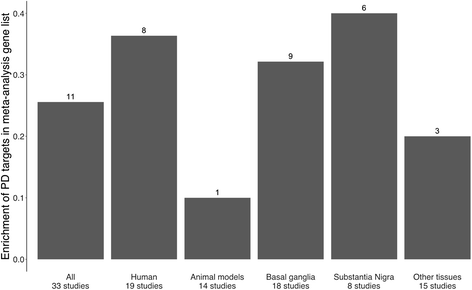
Results: Concordance over all studies is low, with correlation of only 0.05 between differential gene expression signatures on average, but increases within human patients and studies of the same tissue type, rising to 0.38 for studies of human substantia nigra. Agreement of animal models, however, is dependent on model type. Studies of brain tissue from Parkinson's disease patients (specifically the substantia nigra) form a distinct group, showing patterns of differential gene expression noticeably different from that in non-brain tissues and animal models of Parkinson's disease; while comparison with other brain diseases (Alzheimer's disease and brain cancer) suggests that the mixed study types display a general signal of neurodegenerative disease. A meta-analysis of these 33 microarray studies demonstrates the greater ability of studies in humans and highly-affected tissues to identify genes previously known to be associated with Parkinson's disease.
Conclusions: The observed clustering and concordance results suggest the existence of a 'characteristic' signal of Parkinson's disease found in significantly affected human tissues in humans. These results help to account for the consistency (or lack thereof) so far observed in microarray studies of Parkinson's disease, and act as a guide to the selection of transcriptomic studies most representative of the underlying gene expression changes in the human disease.
Keywords: Concordance; Gene expression; Meta-analysis; Microarray; Parkinson’s disease.
Figures
Similar articles
-
Transcriptomic profiles in Parkinson's disease.Exp Biol Med (Maywood). 2021 Mar;246(5):584-595. doi: 10.1177/1535370220967325. Epub 2020 Nov 4. Exp Biol Med (Maywood). 2021. PMID: 33148011 Free PMC article.
-
Correcting Differential Gene Expression Analysis for Cyto-Architectural Alterations in Substantia Nigra of Parkinson's Disease Patients Reveals Known and Potential Novel Disease-Associated Genes and Pathways.Cells. 2022 Jan 7;11(2):198. doi: 10.3390/cells11020198. Cells. 2022. PMID: 35053314 Free PMC article.
-
Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson's disease pathology.Brain. 2009 Jul;132(Pt 7):1795-809. doi: 10.1093/brain/awn323. Epub 2008 Dec 3. Brain. 2009. PMID: 19052140 Free PMC article.
-
Microarrays in Parkinson's disease: a systematic approach.NeuroRx. 2006 Jul;3(3):319-26. doi: 10.1016/j.nurx.2006.05.008. NeuroRx. 2006. PMID: 16815215 Free PMC article. Review.
-
Current status and future directions of gene expression profiling in Parkinson's disease.Neurobiol Dis. 2012 Jan;45(1):76-82. doi: 10.1016/j.nbd.2010.10.022. Epub 2010 Nov 5. Neurobiol Dis. 2012. PMID: 21056669 Free PMC article. Review.
Cited by
-
Transcriptomic Signatures Associated With Regional Cortical Thickness Changes in Parkinson's Disease.Front Neurosci. 2021 Oct 1;15:733501. doi: 10.3389/fnins.2021.733501. eCollection 2021. Front Neurosci. 2021. PMID: 34658772 Free PMC article.
-
Identification of Genes Potentially Associated with Melanoma Tumorigenesis Through Co-Expression Network Analysis.Int J Gen Med. 2021 Nov 19;14:8495-8508. doi: 10.2147/IJGM.S336295. eCollection 2021. Int J Gen Med. 2021. PMID: 34824546 Free PMC article.
-
Rat subthalamic stimulation: Evaluating stimulation-induced dyskinesias, choosing stimulation currents and evaluating the anti-akinetic effect in the cylinder test.MethodsX. 2019 Oct 14;6:2384-2395. doi: 10.1016/j.mex.2019.10.012. eCollection 2019. MethodsX. 2019. PMID: 31681539 Free PMC article.
-
The landscape of multiscale transcriptomic networks and key regulators in Parkinson's disease.Nat Commun. 2019 Nov 20;10(1):5234. doi: 10.1038/s41467-019-13144-y. Nat Commun. 2019. PMID: 31748532 Free PMC article.
-
Transcriptomic signatures of brain regional vulnerability to Parkinson's disease.Commun Biol. 2020 Mar 5;3(1):101. doi: 10.1038/s42003-020-0804-9. Commun Biol. 2020. PMID: 32139796 Free PMC article.
References
-
- Cruz-Monteagudo M, Borges F, Paz-Y-Mino C, Cordeiro MNDS, Rebelo I, Perez-Castillo Y, Helguera AM, Sanchez-Rodriguez A, Tejera E. Efficient and biologically relevant consensus strategy for Parkinson’s disease gene prioritization. BMC Med Genomics. 2016;9:12. doi: 10.1186/s12920-016-0173-x. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

