Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes
- PMID: 28327131
- PMCID: PMC5360028
- DOI: 10.1186/s12915-017-0362-x
Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes
Abstract
Background: Defining the transcriptome and the genetic pathways of pancreatic cells is of great interest for elucidating the molecular attributes of pancreas disorders such as diabetes and cancer. As the function of the different pancreatic cell types has been maintained during vertebrate evolution, the comparison of their transcriptomes across distant vertebrate species is a means to pinpoint genes under strong evolutionary constraints due to their crucial function, which have therefore preserved their selective expression in these pancreatic cell types.
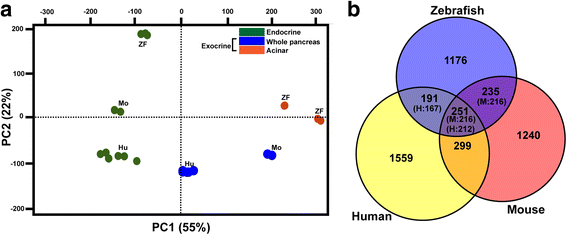
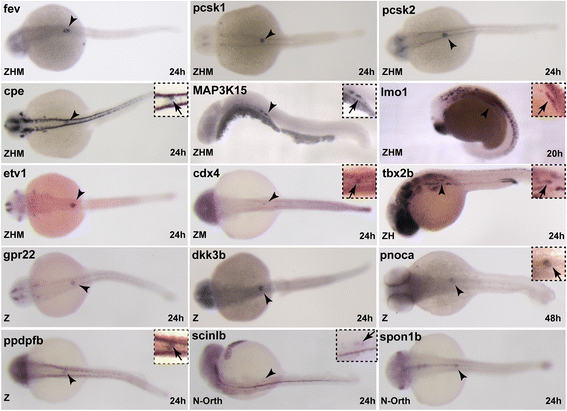
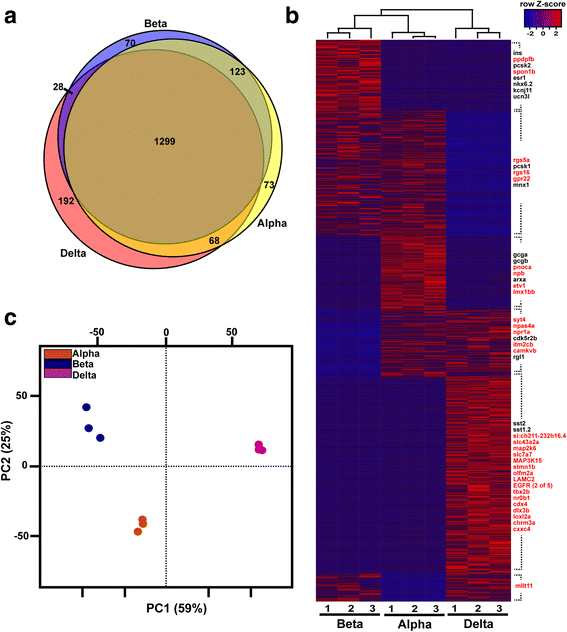
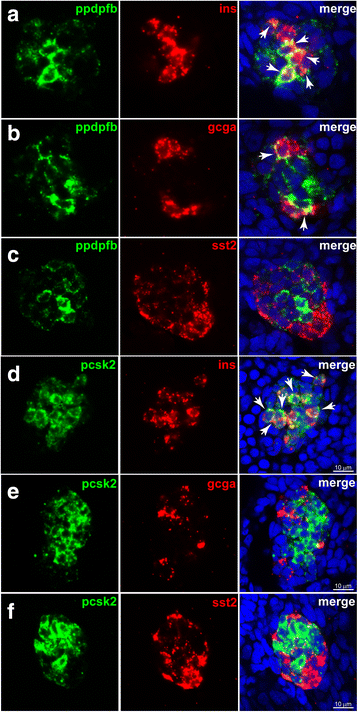
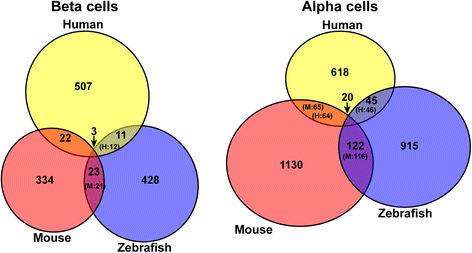
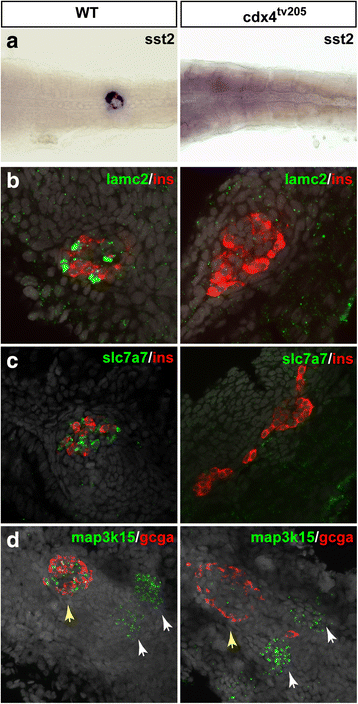
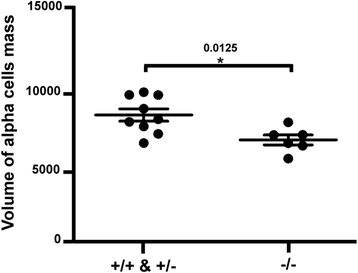
Results: In this study, RNA-sequencing was performed on pancreatic alpha, beta, and delta endocrine cells as well as the acinar and ductal exocrine cells isolated from adult zebrafish transgenic lines. Comparison of these transcriptomes identified many novel markers, including transcription factors and signaling pathway components, specific for each cell type. By performing interspecies comparisons, we identified hundreds of genes with conserved enriched expression in endocrine and exocrine cells among human, mouse, and zebrafish. This list includes many genes known as crucial for pancreatic cell formation or function, but also pinpoints many factors whose pancreatic function is still unknown. A large set of endocrine-enriched genes can already be detected at early developmental stages as revealed by the transcriptomic profiling of embryonic endocrine cells, indicating a potential role in cell differentiation. The actual involvement of conserved endocrine genes in pancreatic cell differentiation was demonstrated in zebrafish for myt1b, whose invalidation leads to a reduction of alpha cells, and for cdx4, selectively expressed in endocrine delta cells and crucial for their specification. Intriguingly, comparison of the endocrine alpha and beta cell subtypes from human, mouse, and zebrafish reveals a much lower conservation of the transcriptomic signatures for these two endocrine cell subtypes compared to the signatures of pan-endocrine and exocrine cells. These data suggest that the identity of the alpha and beta cells relies on a few key factors, corroborating numerous examples of inter-conversion between these two endocrine cell subtypes.
Conclusion: This study highlights both evolutionary conserved and species-specific features that will help to unveil universal and fundamental regulatory pathways as well as pathways specific to human and laboratory animal models such as mouse and zebrafish.
Keywords: Acinar cells; Comparative transcriptomics; Ductal cells; Endocrine cells; Pancreas; RNA-seq.
Figures
Similar articles
-
Transcriptomes of the major human pancreatic cell types.Diabetologia. 2011 Nov;54(11):2832-44. doi: 10.1007/s00125-011-2283-5. Epub 2011 Sep 1. Diabetologia. 2011. PMID: 21882062 Free PMC article.
-
Pax4 is not essential for beta-cell differentiation in zebrafish embryos but modulates alpha-cell generation by repressing arx gene expression.BMC Dev Biol. 2012 Dec 17;12:37. doi: 10.1186/1471-213X-12-37. BMC Dev Biol. 2012. PMID: 23244389 Free PMC article.
-
Progenitor potential of nkx6.1-expressing cells throughout zebrafish life and during beta cell regeneration.BMC Biol. 2015 Sep 2;13:70. doi: 10.1186/s12915-015-0179-4. BMC Biol. 2015. PMID: 26329351 Free PMC article.
-
Acinar cell reprogramming: a clinically important target in pancreatic disease.Epigenomics. 2015;7(2):267-81. doi: 10.2217/epi.14.83. Epigenomics. 2015. PMID: 25942535 Review.
-
Single-Cell RNA-Seq of the Pancreatic Islets--a Promise Not yet Fulfilled?Cell Metab. 2019 Mar 5;29(3):539-544. doi: 10.1016/j.cmet.2018.11.016. Epub 2018 Dec 20. Cell Metab. 2019. PMID: 30581120 Free PMC article. Review.
Cited by
-
Pancreatic gene expression during recovery after pancreatitis reveals unique transcriptome profiles.Sci Rep. 2018 Jan 23;8(1):1406. doi: 10.1038/s41598-018-19392-0. Sci Rep. 2018. PMID: 29362419 Free PMC article.
-
Screening for insulin-independent pathways that modulate glucose homeostasis identifies androgen receptor antagonists.Elife. 2018 Dec 6;7:e42209. doi: 10.7554/eLife.42209. Elife. 2018. PMID: 30520733 Free PMC article.
-
Loss of RREB1 in pancreatic beta cells reduces cellular insulin content and affects endocrine cell gene expression.Diabetologia. 2023 Apr;66(4):674-694. doi: 10.1007/s00125-022-05856-6. Epub 2023 Jan 12. Diabetologia. 2023. PMID: 36633628 Free PMC article.
-
Efficient knock-in method enabling lineage tracing in zebrafish.Life Sci Alliance. 2023 Mar 6;6(5):e202301944. doi: 10.26508/lsa.202301944. Print 2023 May. Life Sci Alliance. 2023. PMID: 36878640 Free PMC article.
-
A whole organism small molecule screen identifies novel regulators of pancreatic endocrine development.Development. 2019 Jul 24;146(14):dev172569. doi: 10.1242/dev.172569. Development. 2019. PMID: 31142539 Free PMC article.
References
-
- Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng Y-H, Roberson RS, Ricordi C, O’Connell PJ, Gonzalez FJ, Kahn CR, O’Connell PJ, Gonzalez FJ, Kahn CR. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–49. doi: 10.1016/j.cell.2005.05.027. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

